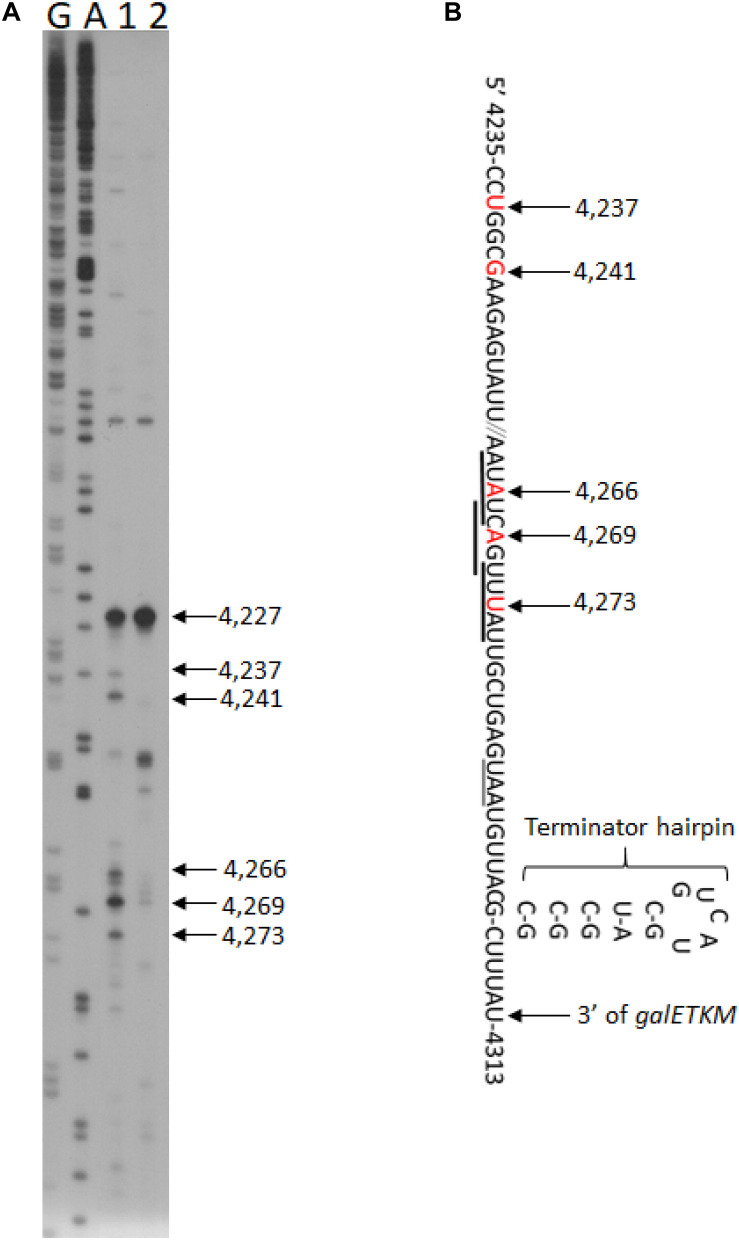
FIGURE 5.
RNase E cleavage upstream of the terminator hairpin of the gal operon. (A) 5′RACE of RNA in front of the terminator hairpin of gal operon isolated from GW20 (ams1ts) cells grown either at the permissive temperature of 30°C (lane 1) or the non-permissive temperature of 44°C (lane 2). The 5′ end at 4,227 is not generated by RNase E cleavage since its intensity did not decrease at the non-permissive temperature. Lanes G and A are DNA sequencing ladders, as in Figure 1E. We note that the 5′ ends located between 4,273 and 4,266 also fall within the consensus sequence for the RNase E cleavage (Belasco, 2017; Chao et al., 2017), but the ends located at 4,241–4,237 seem to deviate from it. Nevertheless, both cleavage clusters would serve the purpose of removing the hairpin, allowing 3′ to 5′ exoribonuclease digestion. (B) Nucleotide sequence of mRNA at the end of galM presented together with their gal coordinates. The nucleotide residue and its gal coordinate that becomes the 5′ end after RNase E cleavage are indicated in red. The RNase E cleavage is indicated with a downward arrow. Matches to the consensus sequence for RNase E cleavage are underlined with thick horizontal lines. The translation stop codon of the galM gene is also underlined (thin line). Also shown is the terminator hairpin. The cytosine residue at 4,313 is the last residue of the full-length mRNA, galETKM (Wang et al., 2019).