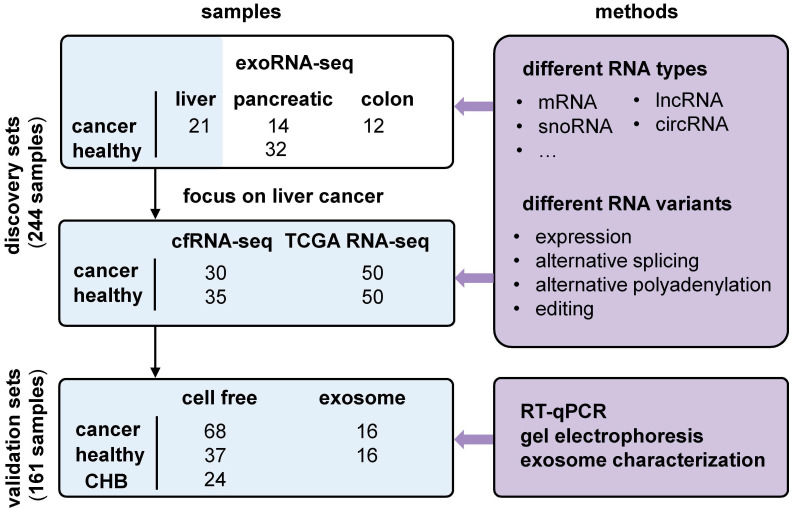
Figure 3.
Overview of experimental design and integrative analysis. Three discovery sets, exosomal RNA-seq (exoRNA-seq) data from exoRBase, self-profiled cell free RNA-seq (cfRNA-seq) data and tissue RNA-seq data from TCGA, were used to discover candidate biomarkers. Two validation sets (qRT-PCR data of cell free and exosomal RNAs) were used for experimental verification. Different RNA types and variations were assayed. Several experimental methods were applied to validate the candidate biomarkers. CHB: chronic hepatitis B.