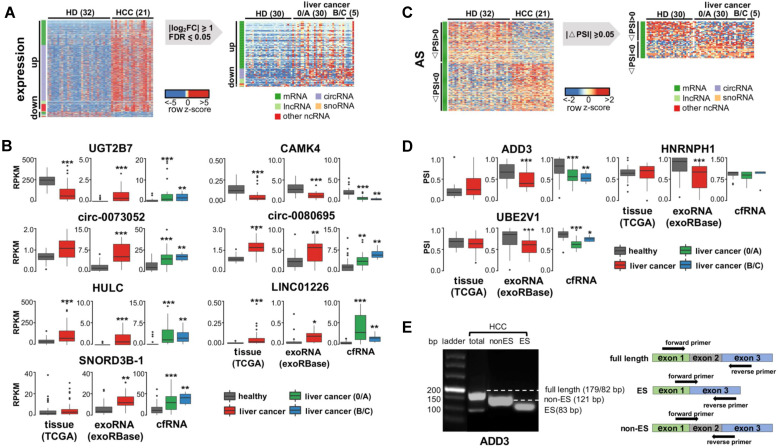
Figure 4.
Identify recurrent exRNA variations from multiple datasets for liver cancer. (A) Differentially expressed exRNAs of liver cancer idenfied from both exoRNA-seq and cfRNA-seq data, using healthy donor as control. (B) Expression level of seven differentially expressed exRNAs in tissue RNA-seq data (TCGA), exoRNA-seq data (exoRBase), and cfRNA-seq data. ***: P-value < 0.001, **: P-value < 0.01, *: P-value < 0.05, Wilcoxon rank sum test. (C) Alternatively spliced exRNAs idenfied from both exoRNA-seq and cfRNA-seq data. (D) Inclusion level of three alternatively spliced exRNAs in tissue RNA-seq data (TCGA), exoRNA-seq data (exoRBase), and cfRNA-seq data. ***: P-value < 0.001, **: P-value < 0.01, *: P-value < 0.05, Wilcoxon rank sum test. (E) Representative gel electrophoresis image showing alternative splicing of ADD3 (left); primers designed for validating alternative splicing (right). Three kinds of primers were designed for full length, exon skipping (ES), and non-exon skipping/exon inclusion (Non-ES).