Abstract
Epigenetic inactivation of GADD45A is a common occurrence in different types of cancer. However, little is known regarding its association with radiosensitivity in cervical cancer (CC). Thus, the present study aimed to investigate the association between aberrant GADD45A methylation and radiosensitivity in CC. SiHa, HeLa and CaSki CC cells were treated with 5-azacytidine (5-azaC), with or without irradiation. The expression levels of GADD45A and AKT related molecules were detected via reverse transcription-quantitative PCR and western blot analyses. The methylation status of GADD45A was assessed via methylation-specific PCR and cell proliferation assays, while clonogenic assays and flow cytometric analysis were performed to assess the function of the genes (GADD45A and AKT) in the CC cell lines. The results demonstrated that methylation of GADD45A was significantly higher in the radioresistant tissues (63.16%) compared with the radiosensitive samples (33.33%). In addition, the surviving fraction of SiHa cells following irradiation with 2 Gy was demonstrated to be highest amongst the three CC cells (CaSki, 57±9.5%; HeLa, 70±4% and SiHa, 75±10%). The survival rate of SiHa cells following treatment with 5-azaC and ionizing radiation (IR) significantly decreased as the radiation dose increased, compared with treatment with IR alone. Following overexpression of GADD45A or treatment with 5-azaC, the radiosensitivity of SiHa cells significantly increased compared with both the control vector and PBS treated groups. In addition, the AKT inhibitor, MK-2206, increased the radiosensitivity of SiHa cells. Notably, aberrant methylation of GADD45A was associated with decreased radiosensitivity in CC, and the PI3K/AKT signaling pathway was essential for radioresistance, which was mediated through downregulation of GADD45A.
Keywords: cervical cancer, radiosensitivity, methylation, GADD45A, PI3K/AKT signaling pathway
Introduction
Despite advancements in the prevention, detection and treatment of diseases during the past decade, cervical cancer (CC) remains the fourth most common malignancy in women worldwide (1). In 2018, the incidence of cervical cancer was 6.6% worldwide (2). Radiotherapy is an effective treatment, particularly for patients with advanced cancer (3). However, patients with advanced stages of CC still suffer from treatment failure due to the development of resistance to radiotherapy (4). Increasing the sensitivity to radiotherapy in patients resistant to it can improve tumor control and decrease the side effects of conventional treatment (5). Technological advancements in radiotherapy, such as intensity-modulated radiotherapy, have contributed to decreased treatment-related toxicity for patients with locally-advanced cancer (6); however, the molecular mechanisms underlying the development of radioresistance, and the associated biomarkers remain unknown. Currently, several gene variations (7), tumor microenvironmental changes (8), hypoxia (9) and specific signaling pathways (10,11) contribute to cellular resistance against radiotherapy. These factors enhance DNA repair, decrease apoptosis and increase genetic instability (7,12). Thus, it remains critical to investigate the molecular mechanism and prognostic markers of radioresistance, in order to increase effectiveness of radiotherapy.
Recent studies have demonstrated an association between the radiosensitivity of cancer cells and methylation levels (13–15). Epigenetic modifications, particularly promoter hypermethylation, which results in silencing of the expression of tumor suppressor genes, contribute to the regulation of cellular events associated with cancer development and progression, such as apoptosis (16), cell cycle (17), proliferation (18) and DNA repair (19); and the mechanisms above have been identified to have an effect on radiosensitivity (20,21).
Previous studies have reported that hypermethylation of DNA is observed in CC (22–25). Currently, few reports have investigated the association between DNA methylation and radioresistance of CC (26,27). Notably, it has been suggested that the GADD45 gene family act as DNA damage-inducing and growth-inhibiting genes, which function as tumor suppressors for targeted therapy (28). Furthermore, it has been demonstrated that GADD45 induces epigenetic inactivation in different types of cancer and cancer cell lines, including anaplastic thyroid cancer, hepatocellular carcinomas, non-Hodgkin, Hodgkin lymphoma, nasopharyngeal, cervical, esophageal and lung carcinoma (28,29).
However, whether aberrant GADD45A methylation is associated with radiosensitivity in CC remains to be determined. Thus, the present study aimed to investigate the function of GADD45A methylation in CC radiotherapy, and determine its underlying molecular mechanism. Taken together, the results of the present study suggest that aberrant GADD45A methylation is associated with decreased radiosensitivity in CC, which is mediated via the PI3K/AKT signaling pathway.
Materials and methods
Cell culture and treatment
The human SiHa, CaSki and HeLa CC cell lines were purchased from The Cell Bank of Type Culture Collection of the Chinese Academy of Sciences. CC cells were plated into 60 mm culture dishes at a density of 6×105 cells/well and maintained in RPMI-1640 supplemented with 10% fetal bovine serum (FBS) and 1% ampicillin and streptomycin (all purchased from Gibco; Thermo Fisher Scientific, Inc.), at 37°C with 5% CO2.
Cells were subsequently treated with 5-azacytidine (5-azaC; Sigma-Aldrich; Merck KGaA) and/or irradiated with 3.6 Gy/min at room temperature using an X-Rad 225 X-ray generator (Precision X-ray Inc., http://precisionxray.com/x-rad/small-animal-irradiator/). The experimental groups were as follows: 5-azaC group (0, 1, 3, 5 or 10 µmol/l for 24, 48 and 72 h); ionizing radiation (IR) group (0, 2, 4, 6, 8, 10 Gy given in a single fraction); PBS group and combined treatment group (IR after pretreatment with 5-azaC for 72 h).
Cell Counting Kit-8 (CCK-8) assay
SiHa CC cells were seeded into 96-well plates at a density of 3×103 and treated with 5-azaC (1, 3, 5 or 10 µmol/l) after 24 h. The CCK-8 assay (Dojindo Molecular Technologies, Inc.) was performed after 24, 48 and 72 h of 5-azaC treatment, and viability was subsequently analyzed at a wavelength of 450 nm using a microplate reader (Thermo Fisher Scientific, Inc.). Cell viability was assessed using the following formula: Cell viability (%) = [(As-Ab)/(Ac-Ab)]x100; where As = absorbance of the experimental well, Ab = blank well absorbance and Ac = control well absorbance.
Clonogenic assay
Cells were seeded into 60 mm dishes at a density of 6×105 cells/well and treated with 5-azaC alone (0 or 5 µmol/l) for 72 h or MK-2206 (0 or 0.5 µmol/l) for 48 h, IR alone (0, 2, 4, 6, 8 or 10 Gy) or IR and 5-azaC/MK-2206. Following treatment, cells were digested, collected via centrifugation (1,000 × g for 5 min at room temperature), diluted and seeded into 6-well plates at different cell densities (100 cells for control, 200 cells for 2 Gy, 800 cells for 4 Gy, 1,800 cells for 6 Gy, 3,000 cells for 8 Gy and 5,000 cells for 10 Gy). After 10–15 days, the cells were fixed in methanol for 15 min and stained with trypan blue solution for 20 min at room temperature, and the number of cell colonies per dish were counted with the naked eye. Colonies containing at least 50 cells were counted. Cell survival curves were constructed using a multi-target single-hit model: S(D)=1-(1-e−D/D0)n; where D is the single dose fraction, and D0 is defined as the given average hit dose per target. The number of sensitive targets in a cell was denoted by n (30). The survival fraction (SF) was determined from the number of colonies formed after treatment relative to the colony counts with the plating efficiency of the non-irradiated cells.
Methylation-specific PCR (MSP)
Genomic DNA was extracted from CC cells (5×106) using a DNA Extraction kit (cat. no. K180001; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. To assess the DNA methylation patterns, an EpiTect® Bisulfite kit (cat. no. 59110; Qiagen, Inc.) was used to perform the sodium bisulfite modification. The primer sequences and annealing temperatures were used as previously described (31). The amplified products were analyzed on 3% agarose gels, stained with ethidium bromide (Thermo Fisher Scientific, Inc.) for 15 min at room temperature and visualized under a UV light.
Reverse transcription-quantitative (RT-q)PCR analysis
Total cellular RNA was extracted using RNAiso Plus (Takara Bio, Inc.). Agarose gel analysis was performed to assess the quality of RNA. cDNA was produced from 1 µg total RNA using the RT Reagent kit (cat. no. RR037A, Takara Bio, Inc.), according to the manufacturer's protocol. qPCR was subsequently performed using the SuperScript III Platinum SYBR Green One-Step qPCR kit (cat. no. 11736059, Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol, and a Bio-Rad CFX96 sequence detection system (Bio-Rad Laboratories Inc.) was used to perform the amplification. The following thermocycling conditions were used: Initial denaturation at 95°C for 5 min, followed by 40 cycles of denaturation at 95°C for 15 sec, and annealing and extension at 60°C for 30 sec. The following primer sequences were used for qPCR: GADD45A forward, 5′-GAGAGCAGAAGACCGAAAGGA-3′ and reverse, 5′-CACAACACCACGTTATCGGG-3′; and GAPDH forward, 5′-ACAACTTTGGTATCGTGGAAGG-3′ and reverse, 5′-GCCATCACGCCACAGTTTC-3′. Relative expression levels were quantified using the 2−ΔΔCq method (32) and normalized to the internal reference gene GAPDH.
Western blotting
SiHa CC cells were homogenized on ice using RIPA lysis buffer (Beyotime Institute of Biotechnology), and centrifuged at 1,200 × g for 5 min at 4°C. The supernatant was collected and stored at −80°C until subsequent experimentation.
Protein concentrations were determined using a BCA Protein assay kit (cat. no. 7780, Cell Signaling Technology, Inc.) and ~40 µg protein/lane was separated via SDS-PAGE on a 10% gel at 120 V constant voltage. The separated proteins were subsequently transferred onto PVDF membranes (EMD Millipore) and blocked with 5% non-fat dry milk (cat. no. 9999, Cell Signaling Technology, Inc.) in PBS with 0.1% Tween-20 for 2 h at room temperature. The membranes were incubated with primary antibodies against GADD45A (1:1,000; cat. no. 4632), AKT (1:1,000; cat. no. 4685), p-AKT (Ser 473) (1:2,000; cat. no. 4060) and GAPDH (1:1,000; cat. no. 5174), overnight at 4°C. All primary antibodies were purchased from Cell Signaling Technology, Inc. Membranes were washed three times with 0.1% TBST, and subsequently incubated with goat anti-rabbit secondary antibody (1:2,000; cat. no. 7074; Cell Signaling Technology, Inc.) at room temperature for 2 h. Protein bands were visualized using SignalFire™ ECL reagent (cat. no. 6883; Cell Signaling Technology, Inc.).
Cell transfection
GFP-GADD45A-expression plasmid (GFP-GADD45A), negative control (GFP-NC) and a constitutively active Akt1-expression plasmid (pT3-myr-AKT-HA) were purchased from Shanghai GenePharma Co., Ltd. and Zhongyuan (https://www.tianyancha.com/brand/b44b6550128), respectively. Briefly, 1 day prior to transfection, SiHa cells were seeded into 60 mm culture dishes at a density of 3×105 until they reached 80% confluence. A total of 4 µg plasmid and 10 µl Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.) were diluted in 200 µl RPMI-1640 without FBS, and set aside for 5 min at room temperature. The solution was subsequently mixed, gently swayed and placed into an incubator for 20 min at room temperature. The transfection complex was directly added to the culture dish, and the transfected cells were obtained after 48 h.
Cell cycle analysis
SiHa cells were directly harvested or treated with MK-2206 (0 or 0.5 µmol/l) for 48 h at room temperature, where the drug was added every 24 h. Cells were washed three times with PBS, and flow cytometric analysis was performed to assess cell cycle distribution. Briefly, when cells reached 70% confluence, they were fixed with 70% ethanol overnight at 4°C. Cells were rehydrated in 5 ml PBS at room temperature for 15 min, collected via centrifugation (at 400 × g for 5 min at room temperature) and resuspended in staining buffer supplemented with 3 µmol/l propidium iodide (cat. no. P1304MP, Thermo Fisher Scientific, Inc.) at room temperature for 30 min. Cells were subsequently incubated with staining buffer for 15 min at room temperature, and cell cycle analysis was performed using a FACScan flow cytometer (Beckman Coulter, Inc.) and CellQuest software version 4.0 (Becton Dickinson).
Tissue samples and grouping criteria
A total of 59 patients with International Federation of Gynecology and Obstetrics 2009 (33) stages IIB-IIIB CC, histologically diagnosed with squamous CC and treated at the Affiliated Tumor Hospital of Guangxi Medical University between September 2015 and June 2016 were enrolled in the present study. The median age of the patients was 45.8 years (age range, 27–74 years). CC tissues were collected from patients who had not received chemotherapy prior to radiation therapy. Patients with standard radiotherapy received concurrent weekly cisplatin (40 mg/m2).
Radioresistant and radiosensitive patients were assessed 6 months after completion of radiation therapy via colposcopically directed biopsy. Patients were considered radiosensitive if the lesions completely subsided at the end of radiotherapy and the tumor could not be observed in the cervix at the 6-month follow-up. Patients were considered radioresistant if there was still a residual tumor visible to the naked eye following completion of radiotherapy, which was confirmed by biopsy in the cervix or the presence of tumor tissue confirmed by cervical biopsy at the 6-month follow-up. The present study was approved by the Research Ethics Committee of Guangxi Medical University (Guangxi, China; approval no. LW2020063), and written informed consent was provided by all participants prior to the study start.
Statistical analysis
Statistical analysis was performed using SPSS version 24.0 (SPSS Inc.). In vitro experiments were performed in triplicate and data are presented as the mean ± standard deviation. Unpaired Student's t-test was used to compare differences between two independent groups, while and one-way analysis of variance, followed by Tukey's post hoc test were performed to compare differences between multiple groups. The χ2 test was used to compare the effect of GADD45A hypermethylation on radiosensitivity in patients with CC. P<0.05 was considered to indicate a statistically significant difference.
Results
Promoter methylation of GADD45A in primary CCs and their association with radioresistance
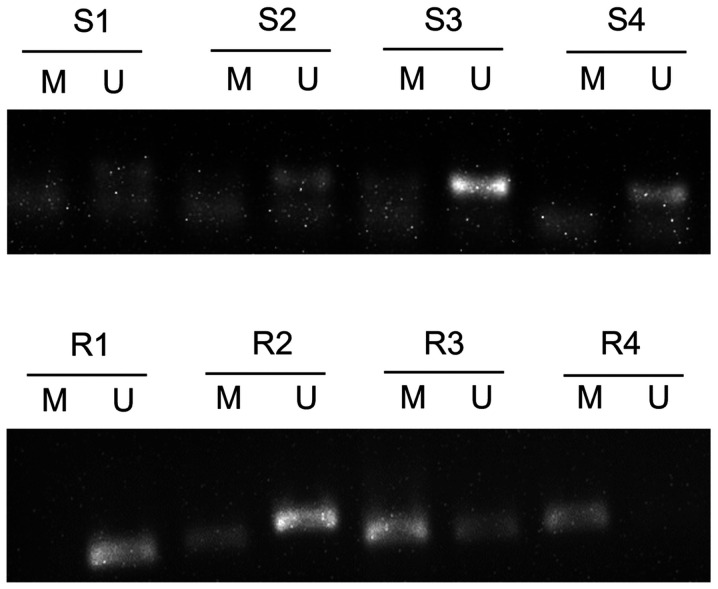
All 59 CC tissue samples were assessed via MSP, and the results demonstrated the possibility of three states: Methylation, incomplete methylation and no methylation (Fig. 1). When incomplete methylation was included into the methylation status for statistical analysis, the results demonstrated that methylation of GADD45A was significantly higher in the radioresistant tissues (63.16%) compared with the radiosensitive samples (33.33%) (P<0.05; Table I). GADD45A promoter methylation in the remaining patients with CC is presented in Fig. S1.
Figure 1.
GADD45A promoter methylation status in patients with CC. GADD45A promoter hypermethylation was higher in the radioresistant tissues compared with the radiosensitive tissues. Promoter methylation of GADD45A in representative CC samples are presented. CC, cervical cancer; R, radioresistant tissues; S, radiosensitive tissues; M, methylated; U, unmethylated.
Table I.
Association between GADD45A hypermethylation and radiosensitivity in cervical cancer tissues.
| Tumor status | GADD45A hypermethylation | P-value |
|---|---|---|
| Radiosensitive | 33.33% (7/21) | <0.05 |
| Radioresistant | 63.16% (24/38) |
Differential GADD45A CpG methylation status, and protein and mRNA levels in the three CC cell lines with different tolerances to radiotherapy
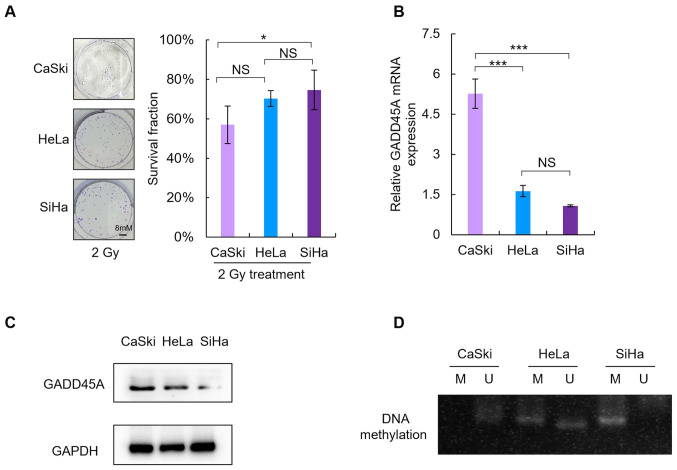
Radiosensitivity was assessed by comparing the SF values in cells treated with 2 Gy irradiation (SF2). As presented in Fig. 2A, the SF2 value of SiHa cells was the highest amongst the three CC cells (CaSki, 57±9.5%; HeLa, 70±4% and SiHa, 75±10%). Consistent with previous findings, the results of the present study demonstrated that SiHa cells exhibited low sensitivity to irradiation, as they had significantly higher SF2 values compared with the other two CC cell lines (P<0.05).
Figure 2.
Differential GADD45A methylation status and radiosensitivity of the three CC cell lines. (A) Radiosensitivity was assessed in CaSki, HeLa and SiHa cells, and determined via a colony formation assay, by measuring the cells as the survival fraction following treatment with 2 Gy radiation. High GADD45A (B) mRNA and (C) protein expression levels were associated with cellular sensitivity to radiation in CC cells. (D) Aberrant methylation of the GADD45A promoter was associated with decreased radiosensitivity in CC cells. *P<0.05, ***P<0.001. CC, cervical cancer; NS, not significant; M, methylated; U, unmethylated.
RT-qPCR and western blot analyses were performed to detect GADD45A mRNA (Fig. 2B) and protein expression levels (Fig. 2C), respectively. Notably, GADD45A mRNA and protein expression levels were lower in SiHa cells compared with the other two CC cell lines.
MSP analysis was performed to determine whether there was an association between the methylation status of GADD45A and downregulation of GADD45A (Fig. 2D). Methylation of GADD45A was evident in SiHa cells; however, a negative association was observed between the methylation status and expression of GADD45A.
Effects of the DNA methylation inhibitor, 5-azaC, on the viability of SiHa cells and the role of GADD45A
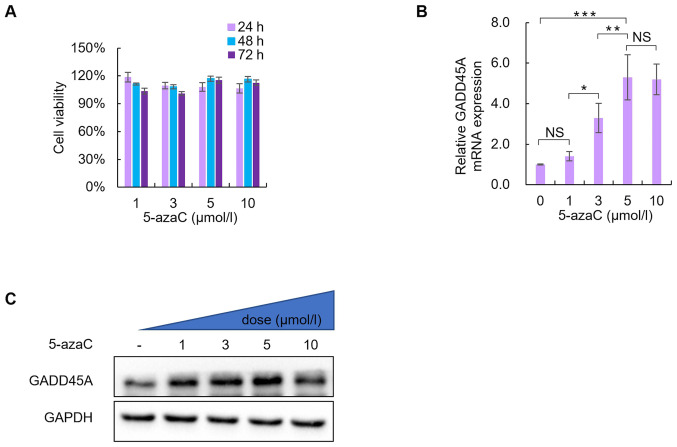
Based on the aforementioned results, SiHa cells were selected for subsequent experimentation. SiHa cells were incubated with 0, 1, 3, 5, or 10 µmol/l 5-azaC to assess the cytotoxic effects of 5-azaC. Cell viability was assessed via the CCK-8 assay following treatment with different concentrations of 5-azaC treatment for 24, 48 and 72 h. As presented in Fig. 3A, no statistically significant differences were observed between treatment with the different concentrations of 5-azaC (P>0.05).
Figure 3.
Effects of the DNA methylation inhibitor, 5-azaC, on the viability of SiHa cells and the role of GADD45A. (A) Effect of 5-azaC treatment on cell viability. No significant change in cell viability was observed following treatment with 1, 3, 5 or 10 µmol/l 5-azaC for 24, 48 and 72 h. GADD45A (B) mRNA and (C) protein expression levels following treatment with 1, 3, 5 or 10 µmol/l 5-azaC for 72 h. PBS was used as the negative control. *P<0.05, **P<0.01, ***P<0.001. 5-azaC, 5-azacytidine; NS, not significant.
Notably, GADD45A mRNA and protein expression levels significantly increased in SiHa cells following treatment with 5 or 10 µmol/l 5-azaC for 72 h (Fig. 3B and C).
5-azaC decreases radioresistance of SiHa cells in vitro
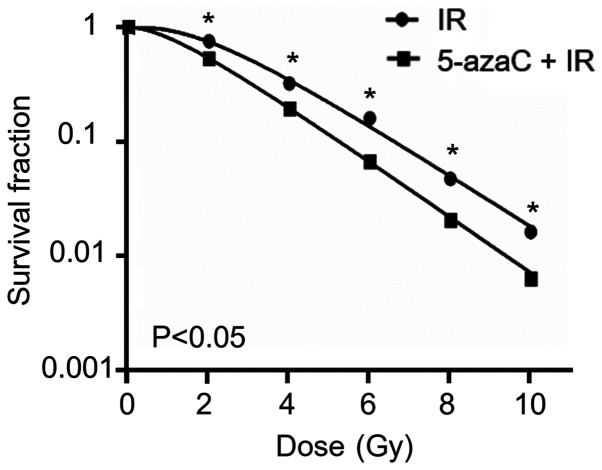
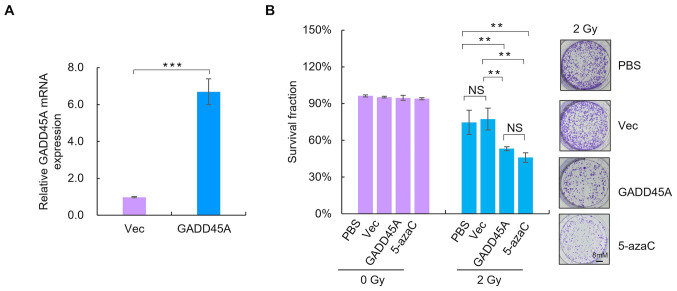
To determine whether the demethylating agent decreased the radioresistance of SiHa cells, the survival fraction of SiHa cells was assessed via the colony formation assay following treatment with 5 µmol/l 5-azaC. The survival fraction of SiHa cells treated with 5-azaC and IR significantly decreased in a dose-dependent manner compared with IR treatment alone (P<0.05; Fig. 4).
Figure 4.
Effect of 5-azaC on the colony formation ability of SiHa cells. Pretreatment with 5 µmol/l 5-azaC for 72 h prior to addition of IR significantly decreased the survival fraction of SiHa cells compared with IR treatment alone. *P<0.05. 5-azaC, 5-azacytidine; IR, ionizing radiation.
Overexpression of GADD45A increases the radiosensitivity of SiHa cells in vitro
To assess the effect of GADD45A on the radiosensitivity of CC cells, GADD45A was overexpressed in SiHa cells using a GFP-GADD45 expression plasmid. RT-qPCR analysis was performed to detect GADD45A mRNA expression levels (Fig. 5A). The clonogenic assay was performed to assess the effect of GADD45A on the sensitivity of SiHa cells to irradiation. As presented in Fig. 5B, overexpression of GADD45A or treatment with the demethylating agent significantly increased the radiosensitivity of SiHa cells compared with the Vec and PBS treated groups, respectively (P<0.05).
Figure 5.
Ectopic GADD45A expression enhances radiosensitivity of SiHa cells. (A) Reverse transcription-quantitative PCR analysis was performed to detect GADD45A mRNA expression 48 h post-transfection. (B) Overexpression of GADD45A decreased the survival fraction of SiHa cells. **P<0.01, ***P<0.001. Vec, vector; NS, not significant.
PI3K/AKT signaling is required for radioresistance, which is mediated by downregulation of GADD45A
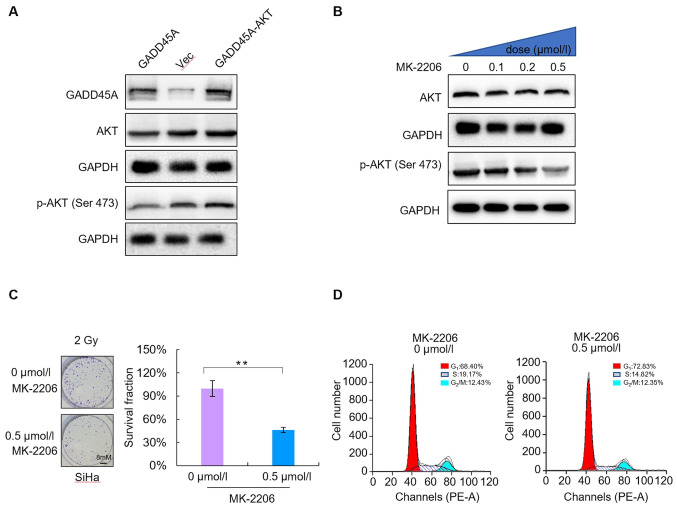
To determine the underlying molecular mechanism of GADD45A-related regulation on the radiosensitivity of CC cells, SiHa cells were transfected with GADD45A and/or AKT DNA. Western blot analysis was performed to detect GADD45A and AKT protein expression levels in SiHa cells. The results demonstrated that co-transfection with GADD45A and AKT reversed the effects of GADD45A overexpression (Fig. 5A). The levels of phosphorylated AKT significantly decreased in a dose-dependent manner following treatment with different concentrations of MK-2206 (0, 0.1, 0.2 and 0.5 µmol/l) for 48 h. In addition, phosphorylation of AKT protein at Ser 473 decreased, whereas total AKT protein levels were not significantly altered (Fig. 6B). The colony formation assay was performed to assess the radiosensitivity of SiHa cells treated with 0.5 µmol/l MK-2206. The results demonstrated that MK-2206 enhanced the radiosensitivity of SiHa CC cells compared with the untreated cells (Fig. 6C). In addition, flow cytometric analysis demonstrated that MK-2206 did not significantly alter the proportion of cells in the different phases of the cell cycle (P>0.05; Fig. 6D). Taken together, these results suggest that the PI3K/AKT signaling pathway is essential for radioresistance, which is mediated by GADD45A downregulation; however, it is not associated with cell cycle distribution.
Figure 6.
PI3K/AKT signaling pathway is essential for radioresistance, which is mediated by downregulation of GADD45A. (A) Phosphorylation levels of AKT were downregulated by GADD45A. (B) MK-2206 inhibited phosphorylation of AKT at Ser 473. (C) MK-2206 enhanced the radiosensitivity of SiHa cells. The colony formation assay was performed to assess the radiosensitivity of SiHa cells. (D) Flow cytometric analysis demonstrated that MK-2206 did not significantly affect the cell cycle distribution. **P<0.01. Vec, vector; NS, not significant.
Discussion
5-azaC is a well-known DNA methyltransferase inhibitor, which induces DNA hypomethylation and epigenetic silencing of gene expression (34). The results of the present study demonstrated that application of a demethylation reagent (5 µmol/l 5-azaC) regulated GADD45A expression. Notably, 5 µmol/l 5-azaC did not exhibit substantial cytotoxic effects in the three CC lines. Furthermore, in vitro experiments indicated that the colony formation ability of SiHa cells was further inhibited by a combination of 5-azaC and IR, compared with IR alone.
Based on current literature, GADD45 family members appear to be infrequently mutated in cancer; however, decreased expression levels of GADD45 family members, due to DNA methylation, have been frequently reported in several types of cancer, including gastric, colorectal and pancreatic cancers (35). The results of the present study demonstrated that GADD45A hypermethylation was significantly higher in the radiotherapy resistance tissues (63.16%) compared with the radiotherapy sensitive tissues (33.33%). Similar results were observed at the cellular level. The GADD45A promoter was completely methylated in SiHa cells, but unmethylated in CaSki cells. Notably, the majority of epigenetic studies in cancer have focused on the methylation of tumor suppressor gene promoters in cell lines, demonstrating different sensitivities (36–38). For example, Kim et al (39) reported that aberrant methylation of ataxia telangiectasia mutated (ATM) is associated with decreased radioresistance in a colorectal cell line, HCT-116.
Based on the effective radiosensitization of 5-azaC, the underlying molecular mechanism was further assessed in SiHa cells. Apoptosis is one of the most common causes of radiosensitization (40). In the present study, 10 µmol/l 5-azaC failed to induce substantial cytotoxicity in unirradiated SiHa cells. However, 5-azaC induced a significant increase in apoptosis during irradiation. In addition, re-expression of GADD45A following treatment with 5-azaC increased the radiosensitivity of SiHa cells. The results demonstrated that the AKT inhibitor, MK-2206, enhanced the radiosensitivity of SiHa cells. It has been reported that the AKT signaling pathway promotes resistance to chemotherapy, radiotherapy and targeted therapy in different types of cancer, including colorectal cancer (41), non-small cell lung cancer (42), ovarian cancer (43) and glioma cell lines (44). Thus, it was hypothesized that downregulation of GADD45A decreases inactivation of the PI3K/AKT signaling pathway, and contributes to cellular resistance against radiotherapy.
Cell cycle arrest is a common cause of increased radiosensitivity (45). In the present study, pretreatment with 5 µmol/l 5-azaC did not impact the cell cycle of SiHa cells. These results are consistent with studies on colorectal cancer (46), nasopharyngeal carcinoma (47), lung cancer (48) and glioblastoma cell lines (49). However, SiHa cells treated with 10 µmol/l 5-azaC remained in the G2/M phase in CC cells (unpublished data). Similar results have been reported following treatment with demethylating agents in breast cancer (50) and endometrial carcinoma (51). The reason for this discrepancy may be due to the different types of cell lines or the different concentrations of demethylation agents used.
In conclusion, the results of the present study demonstrated that hypermethylation of GADD45A was observed in patients with CC, with decreased radiosensitivity, and the PI3K/AKT signaling pathway was required for radioresistance due to downregulation of GADD45A in CC. Taken together, these results suggest that clinical application of epigenetic regulators may be a promising avenue to increase the radiosensitivity of CC.
Supplementary Material
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author upon reasonable request.
Authors' contributions
ML, QZ, and LL designed the present study. ML and LYZ performed the experiments. ML, RL, and TYL analyzed the data. ML and RL drafted the initial manuscript. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by The Research Ethics Committee of Guangxi Medical University (Guangxi, China; approval no. LW2020063), and written informed consent was provided by all participants prior to the study start.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Cohen PA, Jhingran A, Oaknin A, Denny L. Cervical cancer. Lancet. 2019;393:169–182. doi: 10.1016/S0140-6736(18)32470-X. [DOI] [PubMed] [Google Scholar]
- 2.Dickinson JA. Age of initiation of cervical cancer screening. JAMA. 2019;321:611–612. doi: 10.1001/jama.2018.19085. [DOI] [PubMed] [Google Scholar]
- 3.Petrelli F, De Stefani A, Raspagliesi F, Lorusso D, Barni S. Radiotherapy with concurrent cisplatin-based doublet or weekly cisplatin for cervical cancer: A systematic review and meta-analysis. Gynecol Oncol. 2014;134:166–171. doi: 10.1016/j.ygyno.2014.04.049. [DOI] [PubMed] [Google Scholar]
- 4.Global Burden of Disease Cancer Collaboration. Fitzmaurice C, Dicker D, Pain A, Hamavid H, Moradi-Lakeh M, MacIntyre MF, Allen C, Hansen G, Woodbrook R, et al. The Global Burden of Cancer 2013. JAMA Oncol. 2015;1:505–527. doi: 10.1001/jamaoncol.2015.0735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Begg AC, Stewart FA, Vens C. Strategies to improve radiotherapy with targeted drugs. Nat Rev Cancer. 2011;11:239–253. doi: 10.1038/nrc3007. [DOI] [PubMed] [Google Scholar]
- 6.Wang W, Zhang F, Hu K, Hou X. Image-guided, intensity-modulated radiation therapy in definitive radiotherapy for 1,433 patients with cervical cancer. Gynecol Oncol. 2018;151:444–448. doi: 10.1016/j.ygyno.2018.09.024. [DOI] [PubMed] [Google Scholar]
- 7.Ishikawa K, Koyama-Saegusa K, Otsuka Y, Ishikawa A, Kawai S, Yasuda K, Suga T, Michikawa Y, Suzuki M, Iwakawa M, Imai T. Gene expression profile changes correlating with radioresistance in human cell lines. Int J Radiat Oncol Biol Phys. 2006;65:234–245. doi: 10.1016/j.ijrobp.2005.12.048. [DOI] [PubMed] [Google Scholar]
- 8.Theys J, Jutten B, Habets R, Paesmans K, Groot AJ, Lambin P, Wouters BG, Lammering G, Vooijs M. E-Cadherin loss associated with EMT promotes radioresistance in human tumor cells. Radiother Oncol. 2011;99:392–397. doi: 10.1016/j.radonc.2011.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carlson DJ, Yenice KM, Orton CG. Tumor hypoxia is an important mechanism of radioresistance in hypofractionated radiotherapy and must be considered in the treatment planning process. Med Phys. 2011;38:6347–6350. doi: 10.1118/1.3639137. [DOI] [PubMed] [Google Scholar]
- 10.Grana TM, Rusyn EV, Zhou H, Sartor CI, Cox AD. Ras mediates radioresistance through both phosphatidylinositol 3-kinase-dependent and Raf-dependent but mitogen-activated protein kinase/extracellular signal-regulated kinase kinase-independent signaling pathways. Cancer Res. 2002;62:4142–4150. [PubMed] [Google Scholar]
- 11.Skvortsova I, Skvortsov S, Stasyk T, Raju U, Popper BA, Schiestl B, von Guggenberg E, Neher A, Bonn GK, Huber LA, Lukas P. Intracellular signaling pathways regulating radioresistance of human prostate carcinoma cells. Proteomics. 2008;8:4521–4533. doi: 10.1002/pmic.200800113. [DOI] [PubMed] [Google Scholar]
- 12.Bristow RG, Hill RP. Hypoxia and metabolism. Hypoxia, DNA repair and genetic instability. Nat Rev Cancer. 2008;8:180–192. doi: 10.1038/nrc2344. [DOI] [PubMed] [Google Scholar]
- 13.Fabbrizi MR, Warshowsky KE, Zobel CL, Hallahan DE, Sharma GG. Molecular and epigenetic regulatory mechanisms of normal stem cell radiosensitivity. Cell Death Discov. 2018;184:117. doi: 10.1038/s41420-018-0132-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wei W, Dong Z, Gao H, Zhang YY, Shao LH, Jin LL, Lv YH, Zhao G, Shen YN, Jin SZ. MicroRNA-9 enhanced radiosensitivity and its mechanism of DNA methylation in non-small cell lung cancer. Gene. 2019;710:178–185. doi: 10.1016/j.gene.2019.05.050. [DOI] [PubMed] [Google Scholar]
- 15.Sutton LP, Jeffreys SA, Phillips JL, Taberlay PC, Holloway AF, Ambrose M, Joo JE, Young A, Berry R, Skala M, Brettingham-Moore KH. DNA methylation changes following DNA damage in prostate cancer cells. Epigenetics. 2019;14:989–1002. doi: 10.1080/15592294.2019.1629231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hervouet E, Cheray M, Vallette FM, Cartron PF. DNA methylation and apoptosis resistance in cancer cells. Cells. 2013;2:545–573. doi: 10.3390/cells2030545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Desjobert C, El Maï M, Gérard-Hirne T, Guianvarc'h D, Carrier A, Pottier C, Arimondo PB, Riond J. Combined analysis of DNA methylation and cell cycle in cancer cells. Epigenetics. 2015;10:82–91. doi: 10.1080/15592294.2014.995542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pfeifer GP. Defining driver DNA methylation changes in human cancer. Int J Mol Sci. 2018;19:1166. doi: 10.3390/ijms19041166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Russo G, Landi R, Pezone A, Morano A, Zuchegna C, Romano A, Muller MT, Gottesman ME, Porcellini A, Avvedimento EV. DNA damage and repair modify DNA methylation and chromatin domain of the targeted locus: Mechanism of allele methylation polymorphism. Sci Rep. 2016;6:33222. doi: 10.1038/srep33222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Borràs-Fresneda M, Barquinero JF, Gomolka M, Hornhardt S, Rössler U, Armengol G, Barrios L. Differences in DNA repair capacity, cell death and transcriptional response after irradiation between a radiosensitive and a radioresistant cell line. Sci Rep. 2016;6:27043. doi: 10.1038/srep27043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dukaew N, Konishi T, Chairatvit K, Autsavapromporn N, Soonthornchareonnon N, Wongnoppavich A. Enhancement of radiosensitivity by eurycomalactone in human NSCLC cells through G2/M cell cycle arrest and delayed DNA double-strand break repair. Oncol Res. 2020;28:161–175. doi: 10.3727/096504019X15736439848765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feng Q, Hawes SE, Stern JE, Dem A, Sow PS, Dembele B, Toure P, Sova P, Laird PW, Kiviat NB. Promoter hypermethylation of tumor suppressor genes in urine from patients with cervical neoplasia. Cancer Epidemiol Biomarkers Prev. 2007;16:1178–1184. doi: 10.1158/1055-9965.EPI-06-0694. [DOI] [PubMed] [Google Scholar]
- 23.Snoek BC, Splunter APV, Bleeker MCG, Ruiten MCV, Heideman DAM, Rurup WF, Verlaat W, Schotman H, Gent MV, Trommel NEV, Steenbergen RDM. Cervical cancer detection by DNA methylation analysis in urine. Sci Rep. 2019;9:3088. doi: 10.1038/s41598-019-39275-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kazim N, Adhikari A, Oh TJ, Davie J. The transcription elongation factor TCEA3 induces apoptosis in rhabdomyosarcoma. Cell Death Dis. 2020;11:67. doi: 10.1038/s41419-020-2258-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chi HC, Tsai CY, Tsai MM, Lin KH. Impact of DNA and RNA methylation on radiobiology and cancer progression. Int J Mol Sci. 2018;19:555. doi: 10.3390/ijms19020555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiao X, Zhang S, Jiao J, Zhang T, Qu W, Muloye GM, Kong B, Zhang Q, Cui B. Promoter methylation of SEPT9 as a potential biomarker for early detection of cervical cancer and its overexpression predicts radioresistance. Clin Epigenetics. 2019;11:120. doi: 10.1186/s13148-019-0719-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guerrero-Setas D, Pérez-Janices N, Blanco-Fernandez L, Ojer A, Cambra K, Berdasco M, Esteller M, Maria-Ruiz S, Torrea N, Guarch R. LRASSF hypermethylation is present and related to shorter survival in squamous cervical cancer. Mod Pathol. 2013;26:1111–1122. doi: 10.1038/modpathol.2013.32. [DOI] [PubMed] [Google Scholar]
- 28.Zerbini LF, Libermann TA. GADD45 deregulation in cancer: Frequently methylated tumor suppressors and potential therapeutic targets. Clin Cancer Res. 2005;11:6409–6413. doi: 10.1158/1078-0432.CCR-05-1475. [DOI] [PubMed] [Google Scholar]
- 29.Ying J, Srivastava G, Hsieh WS, Gao Z, Murray P, Liao SK, Ambinder R, Tao Q. The stress-responsive gene GADD45G is a functional tumor suppressor, with its response to environmental stresses frequently disrupted epigenetically in multiple tumors. Clin Cancer Res. 2005;11:6442–6449. doi: 10.1158/1078-0432.CCR-05-0267. [DOI] [PubMed] [Google Scholar]
- 30.Zackrisson B. Radiobiological cell survival models. A methodological overview. Acta Oncol. 1992;31:433–441. doi: 10.3109/02841869209088286. [DOI] [PubMed] [Google Scholar]
- 31.Na YK, Lee SM, Hong HS, Kim JB, Park JY, Kim DS. Hypermethylation of growth arrest DNA-damage-inducible gene 45 in non-small cell lung cancer and its relationship with clinicopathologic features. Mol Cells. 2010;30:89–92. doi: 10.1007/s10059-010-0092-1. [DOI] [PubMed] [Google Scholar]
- 32.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 33.Tsikouras P, Zervoudis S, Manav B, Tomara E, Iatrakis G, Romanidis C, Bothou A, Galazios G. Cervical cancer: Screening, diagnosis and staging. J BUON. 2016;21:320–325. [PubMed] [Google Scholar]
- 34.Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457–463. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- 35.Zhang W, Li T, Shao Y, Zhang C, Wu Q, Yang H, Zhang J, Guan M, Yu B, Wan J. Semi-quantitative detection of GADD45-gamma methylation levels in gastric, colorectal and pancreatic cancers using methylation-sensitive high-resolution melting analysis. J Cancer Res Clin Oncol. 2010;136:1267–1273. doi: 10.1007/s00432-010-0777-z. [DOI] [PubMed] [Google Scholar]
- 36.Kim JS, Kim SY, Lee M, Kim SH, Kim SM, Kim EJ. Radioresistance in a human laryngeal squamous cell carcinoma cell line is associated with DNA methylation changes and topoisomerase II α. Cancer Biol Ther. 2015;16:558–566. doi: 10.1080/15384047.2015.1017154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smits KM, Melotte V, Niessen HE, Dubois L, Oberije C, Troost EG, Starmans MH, Boutros PC, Vooijs M, van Engeland M, Lambin P. Epigenetics in radiotherapy: Where are we heading? Radiother Oncol. 2014;111:168–177. doi: 10.1016/j.radonc.2014.05.001. [DOI] [PubMed] [Google Scholar]
- 38.Dote H, Cerna D, Burgan WE, Carter DJ, Cerra MA, Hollingshead MG, Camphausen K, Tofilon PJ. Enhancement of in vitro and in vivo tumor cell radiosensitivity by the DNA methylation inhibitor zebularine. Clin Cancer Res. 2005;11:4571–4579. doi: 10.1158/1078-0432.CCR-05-0050. [DOI] [PubMed] [Google Scholar]
- 39.Kim EH, Park AK, Dong SM, Ahn JH, Park WY. Global analysis of CpG methylation reveals epigenetic control of the radiosensitivity in lung cancer cell lines. Oncogene. 2010;29:4725–4731. doi: 10.1038/onc.2010.223. [DOI] [PubMed] [Google Scholar]
- 40.Muschel RJ, Soto DE, McKenna WG, Bernhard EJ. Radiosensitization and apoptosis. Oncogene. 1998;17:3359–3363. doi: 10.1038/sj.onc.1202580. [DOI] [PubMed] [Google Scholar]
- 41.Yuan YH, Wang HY, Lai Y, Zhong W, Liang WL, Yan FD, Yu Z, Chen JK, Lin Y. Epigenetic inactivation of HOXD10 is associated with human colon cancer via inhibiting the RHOC/AKT/MAPK signaling pathway. Cell Commun Signal. 2019;17:9. doi: 10.1186/s12964-018-0316-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schuurbiers OC, Kaanders JH, van der Heijden HF, Dekhuijzen RP, Oyen WJ, Bussink J. The PI3-K/AKT-pathway and radiation resistance mechanisms in non-small cell lung cancer. J Thorac Oncol. 2009;4:761–767. doi: 10.1097/JTO.0b013e3181a1084f. [DOI] [PubMed] [Google Scholar]
- 43.Zhou HM, Sun QX, Cheng Y. Paeonol enhances the sensitivity of human ovarian cancer cells to radiotherapy-induced apoptosis due to downregulation of the phosphatidylinositol-3-kinase/Akt/phosphatase and tensin homolog pathway and inhibition of vascular endothelial growth factor. Exp Ther Med. 2017;14:3213–3220. doi: 10.3892/etm.2017.4877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Man J, Shoemake JD, Ma T, Rizzo AE, Godley AR, Wu Q, Mohammadi AM, Bao S, Rich JN, Yu JS. Hyperthermia sensitizes glioma stem-like cells to radiation by inhibiting AKT signaling. Cancer Res. 2015;75:1760–1769. doi: 10.1158/0008-5472.CAN-14-3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Otani K, Naito Y, Sakaguchi Y, Seo Y, Takahashi Y, Kikuta J, Ogawa K, Ishii M. Cell-cycle-controlled radiation therapy was effective for treating a murine malignant melanoma cell line in vitro and in vivo. Sci Rep. 2016;6:30689. doi: 10.1038/srep30689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim JG, Bae JH, Kim JA, Heo K, Yang K, Yi JM. Combination effect of epigenetic regulation and ionizing radiation in colorectal cancer cells. PLoS One. 2014;9:e105405. doi: 10.1371/journal.pone.0105405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiang W, Li YQ, Liu N, Sun Y, He QM, Jiang N, Xu YF, Chen L, Ma J. 5-Azacytidine enhances the radiosensitivity of CNE2 and SUNE1 cells in vitro and in vivo possibly by altering DNA methylation. PLoS One. 2014;9:e93273. doi: 10.1371/journal.pone.0093273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Momparler RL. Epigenetic therapy of non-small cell lung cancer using decitabine (5-aza-2′-deoxycytidine) Front Oncol. 2013;3:188. doi: 10.3389/fonc.2013.00188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim HJ, Kim JH, Chie EK, Young PD, Kim IA, Kim IH. DNMT (DNA methyltransferase) inhibitors radiosensitize human cancer cells by suppressing DNA repair activity. Radiat Oncol. 2012;7:39. doi: 10.1186/1748-717X-7-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang L, Zhang Y, Li R, Chen Y, Pan X, Li G, Dai F, Yang J. 5-aza-2′-Deoxycytidine enhances the radiosensitivity of breast cancer cells. Cancer Biother Radiopharm. 2013;28:34–44. doi: 10.1089/cbr.2012.1170. [DOI] [PubMed] [Google Scholar]
- 51.Steiner M, Clark B, Tang JZ, Zhu T, Lobie PE. 14-3-3σ mediates G2-M arrest produced by 5-aza-2′-deoxycytidine and possesses a tumor suppressor role in endometrial carcinoma cells. Gynecol Oncol. 2012;127:231–240. doi: 10.1016/j.ygyno.2012.06.039. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author upon reasonable request.