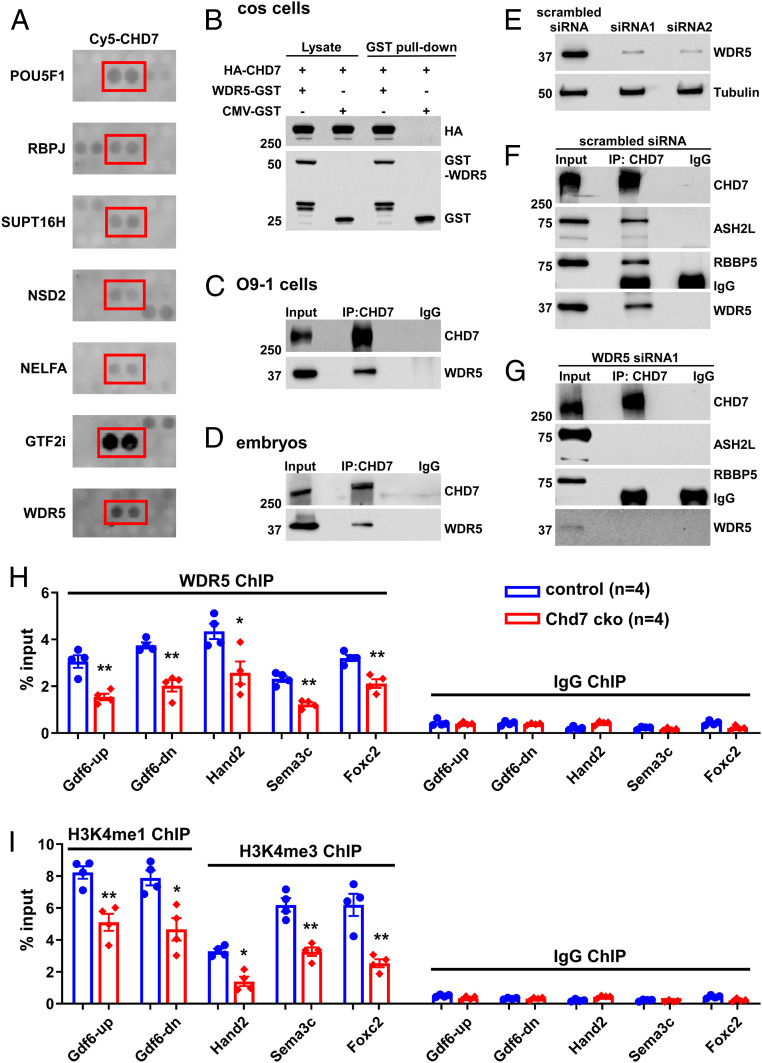
Fig. 5.
CHD7 interacts with and recruits WDR5 to CHD7-target loci. (A) Examples of positive CHD7 interactors from the protein–protein array screen, including known (POU5F1 and RBPJ) and newly identified (including WDR5) direct interactors. (B) COS cells were transfected with different combinations of plasmids encoding HA-CHD7 or WDR5-GST. CMV-GST was included as a control, as indicated on top. Transfections were followed by GST-pull down and Western blot analyses. (C and D) Protein extracts from O9-1 cells (C) or from the trunk region of wild-type embryos at E10.5 (D) were subjected to IP using an anti-CHD7 antibody or a control preimmune IgG. (E–G) Efficient knockdown of WDR5 expression in O9-1 cells (using two independent siRNAs) was assessed by Western blot. Tubulin was used as a protein loading control (E). IP using an anti-CHD7 antibody or a control preimmune IgG were performed using protein extracts from O9-1 cells treated with a scrambled siRNA control (F) or siRNA1 (G). IP of CHD7 and potential co-IP of ASH2L, RBBP5, and WDR5 were assessed by Western blot. The same results were obtained when using siRNA2 (SI Appendix, Fig. S15B). (H and I) ChIP-qPCR was performed using GFP+ cells (cNCCs) isolated from the PA3–6 regions of control (Wnt1-Cre2;Chd7+/+;R26mtmg/+) and mutant (cko, Wnt1-Cre2;Chd7loxp/loxp;R26mtmg/+) embryos at E10.5 to examine association of WDR5 with the indicated promoters/enhancers (H). In addition, H3K4me1 was examined for the enhancers of Gdf6, and H3K4me3 was examined for the promoters of Foxc2, Hand2, and Sema3C (I). *P < 0.05; **P < 0.01, unpaired two-tailed Student’s t test.