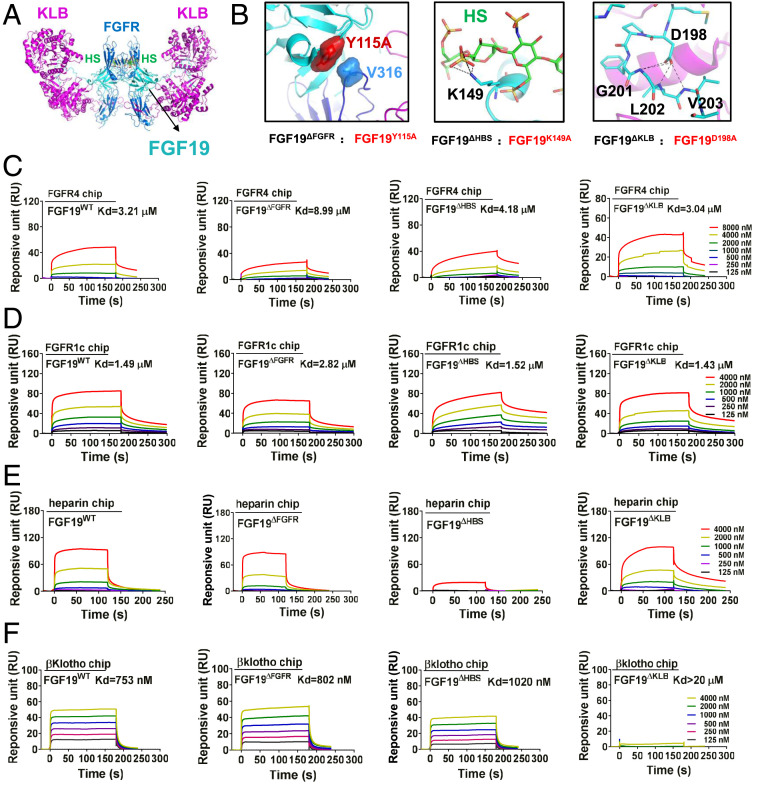
Fig. 1.
Design and characterization of FGF19 variants. (A) A hypothetical 2:2:2:2 FGF19-FGFR1c-KLB-HS dimeric complex was constructed by superimposing structures of FGF19 alone (PDB ID code: 2P23) and of KLB in complex with the C-terminal tail of FGF19 (FGF19CT) (PDB ID code: 6NFJ) onto a dimeric model of a FGF23-FGFR1c-αKlotho-HS dimer (PDB ID code: 5W21). FGF19, FGFR1c, and KLB are shown as cyan, light blue, and purple cartoons, respectively; HS is shown as sticks with carbon, nitrogen, oxygen, and sulfur colored in green, blue, red, and yellow, respectively. (B) Close-up view of interfaces between FGF19 and FGFR (Left), FGF19 and HS (Center), and FGF19 and KLB (Right). In Left, Tyr-115 of FGF19 and Val-316 of FGFR1c are shown as surfaces to highlight their hydrophobic interaction. At Center, the side chain of Lys-149 of FGF19 which engages in hydrogen bonds with HS is shown as sticks. Right shows multiple intramolecular hydrogen bonds mediated by Asp-198 that facilitate FGF19CT conformation, thereby contributing to KLB binding. In Center and Right, black dashed lines denote hydrogen bonds. (C–F) Representative SPR sensorgrams of binding of FGF19WT and FGF19 variants to the ligand-binding domains of FGFR4 (C) and FGFR1 (D), and to HS (E) and KLB (F). Equilibrium dissociation constants (Kd values) were derived from saturation binding curves.