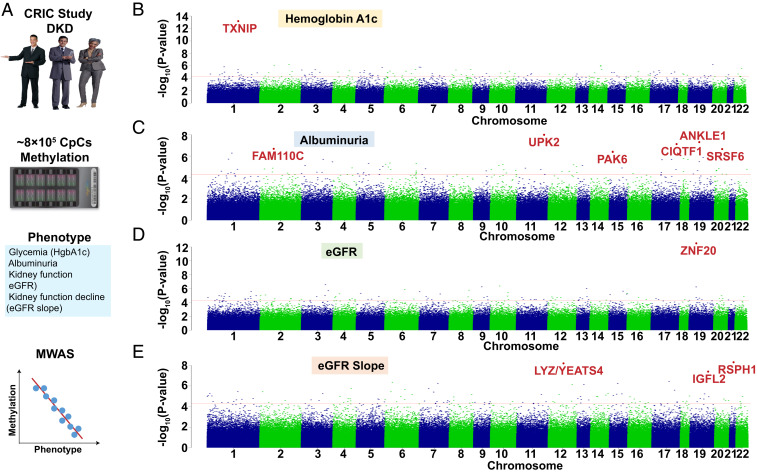
Fig. 1.
Methylation changes associated with DKD phenotypes (glycemia, albuminuria, eGFR, and eGFR slope) in whole-blood samples of the CRIC study participants. (A) Study schematics. Methylation levels of ∼800,000 loci measured by Illumina Human MethylationEPIC arrays were used to analyze the associations with DKD phenotypes (glycemia, albuminuria, eGFR, and eGFR slope) using a linear regression model. (B) Manhattan plot. The y axis is the −log10 of the association P value of hemoglobin A1c and methylation. The x axis represents the genomic location of the CpG probes. n = 473 samples. (C) Manhattan plot. The y axis is the −log10 of P value of albuminuria (24 h) and methylation. The x axis represents the genomic location of the CpG probes. n = 473 samples. (D) Manhattan plot. The y axis is the −log10 of P value of kidney function (baseline eGFR) and methylation. The x axis represents the genomic location of the CpG probes. n = 473 samples. (E) Manhattan plot. The y axis is the −log10 of P value of kidney function decline (eGFR slope) and methylation. The x axis represents the genomic location of the CpG probes. n = 410 samples.