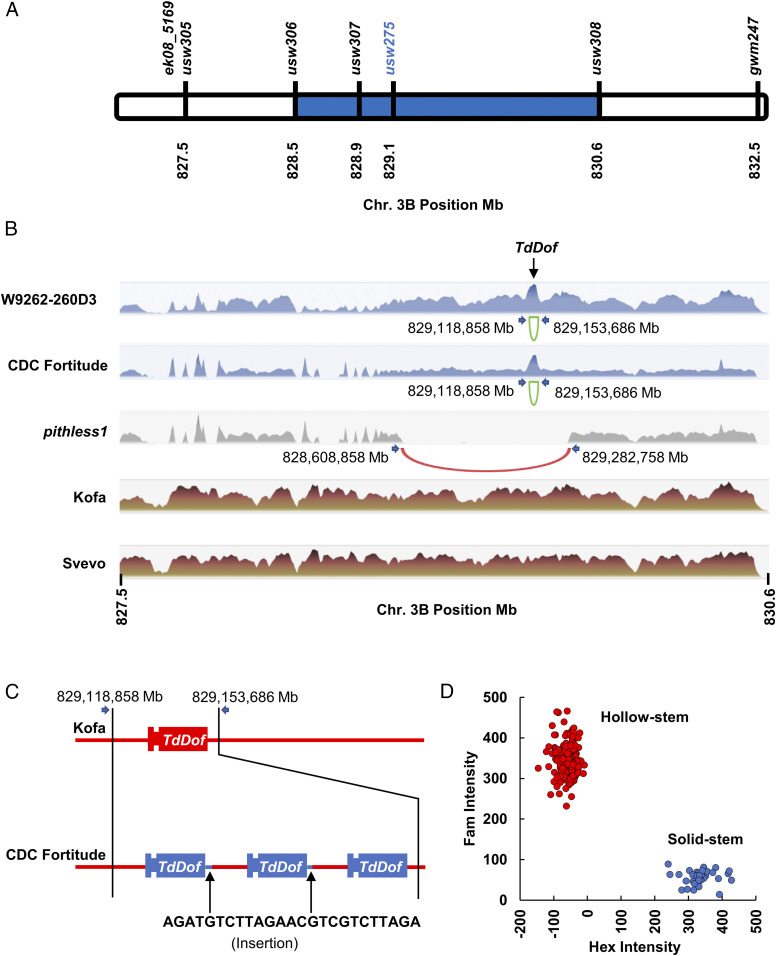
Fig. 1.
CNV of TdDof is associated with stem solidness in wheat. (A) Fine-map interval of SSt1 in the Kofa/W9262-260D3 F2 population. The region between closest flanking markers usw306 and usw308 is shown in blue. (B) Chromium read coverage plots within the SSt1 interval on chromosome 3B of five durum lines. The y axis of each coverage plot denotes the minimum and maximum read coverage for each line within the interval. GemCode molecule associations spanning structural variants are indicated by the green bars (CDC Fortitude, W9262-260D3) and red bar (pithless1). Positions are in megabases. (C) Diagram showing the organization of the TdDof CNV region in CDC Fortitude (solid-stemmed) and Kofa (hollow-stemmed) with CNV breakpoints indicated by the black bars. Three copies of TdDof (TdDof1-3) are present in CDC Fortitude vs. a single copy in Kofa. At the CNV breakpoint between TdDof1-2 and TdDof2-3, a unique 25-bp insertion is present that is only found in solid-stemmed cultivars. (D) High-throughput KASP (kompetitive allele-specific PCR) marker usw275 accurately distinguishes stems that are hollow (red) and solid (blue) in both bread and durum wheat.