Significance
Pheno-RNA is a new idea/method to identify genes important for a phenotype. It involves 1) generating a phenotypic series that involves different experimental conditions to yield a measurable phenotype, 2) performing transcriptional profiling under all the conditions, and 3) correlating gene expression profiles with phenotypic strength. Using this method, we identified ∼200 genes whose expression profiles over 17 conditions show a remarkably high correlation to the level of transformation. As expected, these ∼200 genes are enriched in biological categories important for transformation. Within these categories, some genes have very high correlations with the transformation phenotype, whereas others do not. Ninety genes with high correlations have not been previously linked to cancer, suggesting heretofore unknown genes with a role in cancer.
Keywords: cancer, cellular transformation, mRNA profiling, phenotypic analysis
Abstract
Cellular transformation is associated with dramatic changes in gene expression, but it is difficult to determine which regulated genes are oncogenically relevant. Here we describe Pheno-RNA, a general approach to identifying candidate genes associated with a specific phenotype. Specifically, we generate a “phenotypic series” by treating a nontransformed breast cell line with a wide variety of molecules that induce cellular transformation to various extents. By performing transcriptional profiling across this phenotypic series, the expression profile of every gene can be correlated with the strength of the transformed phenotype. We identify ∼200 genes whose expression profiles are very highly correlated with the transformation phenotype, strongly suggesting their importance in transformation. Within biological categories linked to cancer, some genes show high correlations with the transformed phenotype, but others do not. Many genes whose expression profiles are highly correlated with transformation have never been associated with cancer, suggesting the involvement of heretofore unknown genes in cancer.
A variety of whole-genome approaches have been used to determine which genes are important for a specific phenotype. Transcriptional profiling of cells under the relevant developmental or environmental conditions identifies differentially expressed genes (1–4). While some of these are likely important for the process of interest, it is unlikely that all of them are. In this regard, there is surprisingly limited conservation of genes in different yeast species that respond to a given stress (5). Alternatively, classical or systematic genome-scale mutational screens identify genes that affect a given phenotype (6–8). However, mutations might confer such phenotypes by indirect and/or artifactual effects, and many important genes might be missed owing to the nature of the assay or to functional redundancy. Integration of other forms of information (e.g., genome binding, chromatin status, protein–protein interactions, genetic epistasis, evolutionary conservation) is very helpful in addressing the connection between genes and phenotypes (9). Here we describe a new genome-scale approach, termed Pheno-RNA, to identifying genes associated with a specific phenotype.
The key feature of Pheno-RNA is a phenotypic series, a set of quantitatively measured phenotypes ranging from null to severe generated by treating cells with a wide variety of experimental perturbations. The specific perturbations, individually or in combination, do not matter for Pheno-RNA analysis as long as they generate a series of phenotypes from weak to strong. The expression profiles of individual genes under these various experimental conditions are then correlated with a quantitative measurement of the phenotype. In principle, the expression profiles of genes driving or otherwise relevant for the phenotype should be highly correlated with the degree/strength of the phenotype. Conversely, genes whose expression is regulated by only one or a few experimental conditions are likely passengers under those conditions and not generally relevant for the phenotype. Thus, unlike sophisticated approaches that use multiple transcriptional profiles to correlate gene expression to the presence or absence of a phenotype (10–16), Pheno-RNA correlates the transcriptional profile of an individual gene to the strength of the phenotype.
We apply Pheno-RNA to the process of cellular transformation, using an inducible model in which transient activation of v-Src oncoprotein converts a nontransformed breast epithelial cell line into a stably transformed state within 24 h (10, 17). This epigenetic switch between stable nontransformed and transformed states is mediated by an inflammatory positive feedback loop involving NF-κB, STAT3, and AP-1 factors (17, 18), and many genes are directly and jointly coregulated by these factors (19). In addition, we identified >40 transcription factors important for transformation in this model, as well as putative target sites directly bound by these factors (20). The transcriptional regulatory circuits that are important for transformation in this model are also used in cancer cell lines and human cancers from diverse developmental lineages, and they are the basis of an inflammation index that types human cancers by functional criteria (20). Nevertheless, and despite the molecular description of these regulatory circuits, it is unclear which genes are important for transformation. Here we identify >200 genes whose expression profiles are highly correlated with the degree of transformation, 90 of which have not previously been associated with cancer.
Results
Creating a Phenotypic Series.
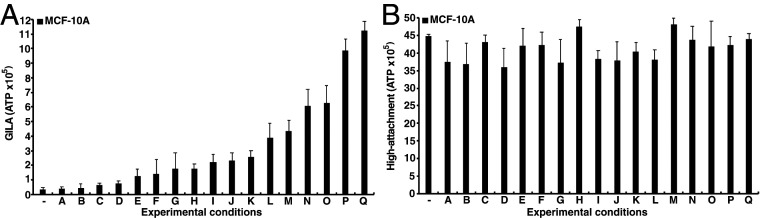
To identify genes associated with breast cellular transformation, we generated a series of transformation phenotypes by treating the parental MCF-10A cell line (i.e., lacking the ER-Src derivative) with a wide variety of signaling molecules—interleukins, growth factors (RGF, FGFs, HGF, TGFs), TNF-⍺, IFN-γ, leptin, insulin, TPA, cAMP—either individually or in combination. The degree of transformation was determined quantitively by growth in low attachment, an assay highly correlated with the more qualitative soft agar assay (21). With the exception of Oncostatin M, none of signaling molecules tested individually caused transformation. Out of hundreds of combinations tested (Dataset S1), we selected 17 combinations to generate the phenotypic series. These 17 treatments show a wide range of transformation phenotypes from very mild to severe (Fig. 1A). Importantly, these treatments do not significantly affect cell growth in standard (high-attachment) conditions (Fig. 1B).
Fig. 1.
Transformation levels and differentially expressed genes. Relative growth levels of MCF-10A cells grown on low-attachment (A) or high-attachment (B) plates for 5 d in 17 different conditions (A to Q and see Dataset S1): A, LPS; B, TGF-⍺ + IFN-Y; C, IFN-Y; D, TGF-⍺ + HGF + IFN-Y; E, HGF + IFN-Y; F, LIGHT; G, TGF-⍺ + HGF; H, Onco M + IFN-Y; I, Onco M+ TGF-⍺ + IFN-Y; J, Onco M + TGF-⍺ + HGF + IFN-Y; K, Onco M + LPS; L, Onco M + HGF + IFN-Y; M, Onco M; N, Onco M + LPS+ LIGHT; O, Onco M + TGF-⍺; P, Onco M + LIGHT; Q, Onco M + TGF-⍺ + HGF; and (−) as control.
Transcriptional Profiling across the Phenotypic Series.
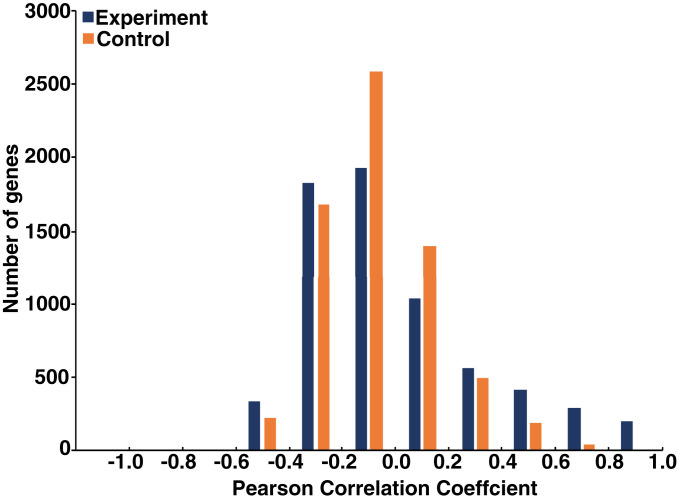
We performed RNA-seq experiments to obtain transcriptional profiles under these 17 conditions (Dataset S2) and analyzed 6,830 genes that gave measurable values under all conditions. As expected, a few thousand genes were up- or down-regulated (by more than twofold) in each condition compared with the untreated control (Fig. 2A), and some genes were up- or down-regulated under multiple conditions (range, 0 to 17; Fig. 2B). We then determined the Pearson’s correlation coefficient between the treated:untreated ratios of mRNA levels and the degree of transformation under the 17 experimental conditions (Dataset S3). The correlation coefficients range from 0.99 to −0.60, compared with the ±0.4 range observed when mRNA values are randomized among the different conditions (Fig. 3). Some genes have remarkably high positive correlation coefficients (209 with >0.8 and 82 with >0.9) that are not observed for any gene in the randomized control data, strongly suggesting their relevance for cellular transformation.
Fig. 2.
Differentially expressed genes. (A) Number of genes with expression induced (black) or reduced (gray) by twofold under the 17 conditions. The control represents the absence of any compound added to the cells. (B) Number of genes with expression induced or reduced under the indicated number (0 to 17) of experimental conditions.
Fig. 3.
Correlation of gene expression profiles to transformation phenotype. Number of genes in the indicated bins of correlation coefficients in the experimental (purple) and shuffled (orange) RNA samples.
The results for negatively regulated genes are less dramatic, but 52 genes have a correlation coefficient <−0.5. Importantly, for any individual condition, only a subset of genes (range 0.5 to 5.5%) showing regulated expression (Fig. 2A) have a high correlation (>0.6; Fig. 3) with the transformation phenotype.
Genes Highly Correlated with Transformation Are Enriched in Expected Cancer-Associated Pathways.
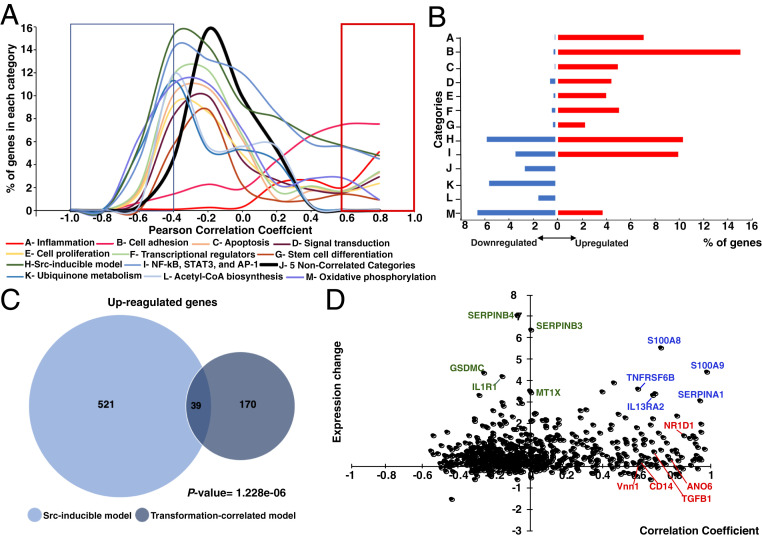
As expected, genes whose expression profiles are highly correlated (either positively or negatively) with transformation are enriched in pathways strongly associated with cancer progress, such as the inflammatory response, apoptosis, metastasis, and transcriptional regulators (Fig. 4 A and B and Dataset S4). In addition, genes showing strong correlation (>0.8) with the transformation phenotype are enriched for genes that are up-regulated in the Src-inducible model (Fig. 4C) (19, 20). However, 81% of these high-correlated genes are not regulated in the Src-inducible model of transformation. Thus, Pheno-RNA confirms genes and pathways previously linked to the transcriptional phenotype and also identifies genes not identified by other approaches.
Fig. 4.
Functional categories of genes. (A) Percentage of genes in each functional category (A to M, shown in various colors) in the indicated bins of correlation coefficients. The red box indicates high positive correlation (>0.6), and the blue box indicates high negative correlation (<−0.4). (B) Percentage of genes in the indicated category (A to M as above) with high positive (>0.6; red) or negative (<−0.5; blue) correlation to the level of transformation. (C) Venn diagram showing the relationships of genes with correlation coefficients >0.8 and those induced by more than twofold by Src induction, including P-values. (D) Relationships between the correlation coefficients and fold induction on Src induction (log2) for all genes (black dots) that are part of the NF-κB/STAT3/AP-1 transcriptional network. Selected inflammatory genes with high correlation/high induction (blue), low correlation/high induction (green), and high correlation/low induction (red) are indicated.
Distinguishing among Genes within a Cancer-Associated Pathway That Are or Are Not Strongly Correlated with the Transformation Phenotype.
Although the inflammatory pathway is critical for transformation, our results (Fig. 4D) distinguish among inflammatory genes whose expression patterns are strongly correlated with the transformation phenotype (examples in red and blue) from those that are not (examples in green). Some inflammatory genes that are induced by Src have expression levels highly correlated with the transformation phenotype (examples in blue), whereas other Src-inducible genes show essentially no correlation with the transformation phenotype (examples in green). Conversely, other inflammatory genes have expression profiles highly correlated with transformation but are not induced in our ER-Src model (examples in red). These results suggest the presence of a subset of inflammatory genes important for transformation. Similar analyses of noninflammatory pathways previously linked to transformation (e.g., apoptosis, cell proliferation, metastasis) also reveal subsets of genes whose expression patterns strongly correlate with transformation.
Identification of Genes Highly Correlated to the Transformation Phenotype That Have Not Been Previously Associated with Cancer.
As Pheno-RNA is based solely on the connection between specific phenotypes and gene expression patterns in a defined experimental system, it provides an unbiased approach to identifying genes associated with transformation. Therefore, we used the following approach to identify genes whose expression patterns are strongly correlated to the transformation phenotype but have never been previously associated with cancer. First, we performed an automated PubMed search (National Cancer for Biotechnology Information’s E-Utilities) on each gene ID and asked whether it was linked to studies on PubMed. Second, we reviewed the literature to confirm that the candidate cancer genes were not associated with cancer. Third, we cross-checked the candidate genes against the Network of Cancer Genes, which contains detailed information on 2,372 cancer genes; the Cancer Gene Census database, which catalogs genes with mutations causally implicated in cancer; Tumor-Associated Genes; and COSMIC and cBIOPortal, which focus on cancer alterations rather than on cancer genes.
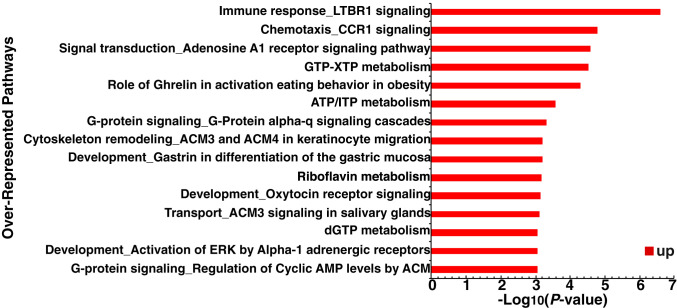
Using this approach, we identified 90 genes strongly linked to the transformation phenotype (R >0.8) but not previously associated with cancer (Dataset S3). Interestingly, these genes are enriched in biological pathways, including immune response LTBR1, chemotaxis CCR1, and adenosine A1 receptor signaling pathways (Fig. 5). Some of these 90 genes are up-regulated in the Src-inducible model, whereas others are not. Conversely, we identified six genes negatively linked to the transformation phenotype (R <−0.5) and previously not associated with cancer. These genes are new candidates for a role in cancer and thus worthy of functional analysis, either individually or in combination.
Fig. 5.
Overrepresented gene categories of 90 genes with high correlation (>0.8) to the transformed phenotype but no previous association with cancer. The significance of each category is indicated by −log10(P-value).
Discussion
Pheno-RNA, a Method to Identify Genes Strongly Associated with a Phenotype.
Our Pheno-RNA analysis distinguishes genes whose expression profiles are strongly associated with the degree of cellular transformation as opposed to genes that are regulated by a specific condition (e.g., Src induction) that causes a transformation phenotype. It also identifies new genes that are candidates for affecting the phenotype. Presumably, genes with high correlation coefficients are important for the transformation phenotype assayed here (i.e., growth under conditions of low attachment). Conversely, regulated genes with poor correlation coefficients are likely to be more relevant for nononcogenic function(s) of the specific experimental condition(s). However, it remains possible that some of these low-correlating genes are important for the transformation phenotype, particularly if pairwise (or higher-order) interactions between genes affect their transcriptional profiles and oncogenic function.
In principle, genes with high correlation coefficients could either drive the phenotype or be coregulated with a pathway important for the phenotype. In this regard, the 90 genes with high correlation coefficients that were not previously associated with cancer nevertheless are enriched in biological pathways (e.g., immune response, CCR signaling, adenosine A1 receptor signaling) associated with cancer. As such, Pheno-RNA can help identify previously unrecognized genes within a pathway that contribute to a phenotype. In addition, it provides independent evidence for the importance of the pathway in mediating the phenotype.
More generally, Pheno-RNA can be applied to multiple phenotypes generated by the experimental conditions. For example, the experimental conditions and transcriptional profiling data presented here can be linked to other transformation-related phenotypes, such as morphology, focus formation, cell motility/invasive growth, mammosphere formation, and metformin sensitivity. The phenotypic series will likely differ among these various phenotypes, making it possible to identify different sets of genes associated with these phenotypes. Finally, Pheno-RNA should be generally applicable for any phenotype that can be quantitated and for which multiple experimental conditions can be used to generate a phenotypic series.
Implications for Cellular Transformation.
Our Pheno-RNA analysis identified 208 genes whose expression profiles are highly correlated (>0.8) with the degree of cellular transformation. This high correlation is far beyond chance expectation (as defined by randomized shuffled data, which revealed no genes with a correlation >0.8), suggesting the relevance of these genes for cellular transformation. Only some of these genes are induced by Src in the original model system, thereby helping distinguish transformation-related genes from those induced by Src for nononcogenic reasons. Conversely, some genes showing very high correlation coefficients are poorly induced by Src activation. Thus, Pheno-RNA provides a more robust list of transformation-associated genes than previously obtained from the ER-Src model.
As expected, the transformation-associated genes identified by Pheno-RNA are enriched in biological categories linked to cancer. Importantly, Pheno-RNA can help discriminate the functional importance of genes within these cancer-related categories, because some genes show very strong correlations, whereas others do not. In addition, we found 90 genes with very high correlations that have not been previously associated with cancer by various approaches. These 90 genes are enriched in certain biological categories, some of which are known to be meaningful for transformation. As such, Pheno-RNA can identify genes within a cancer-related pathway that were not previously associated with cancer. Of course, genetic validation is required to prove functional importance for any individual gene, and gene and/or pathway redundancy might require simultaneous depletion of multiple proteins to observe effects on transformation efficiency. Nevertheless, our results provide a prioritized list of genes to be examined for their effect on cellular transformation, and it seems likely that some of them have a functional role.
Materials and Methods
Cell Culture.
The nontransformed breast cell line MCF-10A (American Type Culture Collection) (22) was cultured in DMEM/F-12 (Thermo Fisher Scientific) supplemented with 5% FBS, 20 ng/mL epidermal growth factor, 0.01 mg/mL insulin, 500 ng/mL hydrocortisone, 100 ng/mL cholera toxin, and 1% antibiotics (penicillin/streptomycin) in a 5% CO2 humidified incubator at 37 °C (10, 17). Cells were subjected to no more than eight passages in culture before their use in experiments. Treatment was performed with different combinations of the factors listed in Dataset S1 at the annotated concentration or with equal volumes of PBS. All growth factors and molecules were purchased from Sigma-Aldrich.
Transformation Assay.
The levels of transformation mediated by the compounds listed in Dataset S1 were determined by a growth in low attachment (GILA) assay essentially as described previously (21), except that 384-well plates were used and the cell concentration was optimized to 25 cells per well in 30 μL. As a control for cell growth, the same samples were assayed on standard plates that permit high attachment. Cells were assayed for ATP content as a surrogate for number of viable cells after 5 d of incubation at 37 °C. Each plate assay had five replications, and the entire screen was repeated three times. The data are presented as the average of these replicates. Cell viability was measured with the CellTiter-Glo (Promega) luminescent assay using the EnVision plate reader (PerkinElmer).
Transcriptional Profiling.
Transcriptome-level mRNA profiling was performed by 3′READS as described previously (23). Total RNA was extracted with TRIzol (Invitrogen) and purified using QIAGEN RNeasy columns with DNase I treatment according to the manufacturer’s instructions. Barcoded RNA-seq libraries were constructed using the 3′READS procedure, and the amplified libraries were purified with SizeSelector-I beads (Aline Biosciences) as described previously (23). Purified barcoded libraries were then quantified with a Bioanalyzer (Agilent) and sequenced at the Harvard Bauer Core Facility using the Illumina NextSeq 500 system. Raw reads were aligned to GENCODE-defined transcripts and the human reference genome (hg19) using Bowtie, allowing one mismatch. The number of mapped reads ranged between 6 and 22 million among the samples. For each condition, read counts for each gene were normalized to the total number of true Poly(A) read pairs, and gene expression levels were indicated as reads per million (RPM) values.
The analyses were performed on the 6,830 genes that gave a measurable signal in all 17 conditions as well as in the control condition. Differentially expressed genes under a given condition (Fig. 2 A and B) were identified by comparing their normalized gene expression values in parallel cultures of treated cells vs. untreated cells. A differentially expressed gene was defined arbitrarily as having a greater than twofold difference, either positive or negative, between treated and untreated cells. As a control, untreated cells were mock-treated with medium lacking inducers; the small set of genes showing a greater than twofold difference represent experimental variation. As shown in Fig. 2A, the number of differentially expressed genes in an experimental condition is approximately 10-fold greater than in the control experiment, indicating that the false discovery rate (FDR) for each condition is ∼10%.
Correlation between Transformation Levels and Gene Expression Profiles.
For all experimental conditions, normalized read counts for each of the 6,830 expressed genes (Dataset S2) were correlated to the level of transformation (ATP values in cells grown in low attachment) by calculating Pearson correlation coefficients (r) using the Excel CORREL function. As a control, RNA values for every gene were randomly shuffled across the 17 treatments and then correlated to the transformation level. Correlation coefficients >0.8 are highly significant, as such values were never obtained in five independent random shuffling experiments. For correlation coefficients between 0.6 and 0.8, 299 were observed with experimental data, as opposed to only 25 with the shuffled data, yielding an FDR of 8.4% for this bin. The overrepresentational significance (P value) for each set of genes was calculated using the Fisher exact test.
Gene Ontology Analyses.
Gene sets based on correlation cutoffs were used as input for MetaCore Functional Enrichment by Gene Ontology using pathway maps to identify biological pathways associated with transformation (https://portal.genego.com/cgi/data_manager.cgi). An FDR cutoff of 0.05 was used to identify pathway maps significantly associated transformation. Network analysis were also performed using Gene Set Enrichment Analysis (GSEA) and DAVID ontology.
Supplementary Material
Acknowledgments
We thank Asaf Rotem for help with the GILA assays and discussions on cancer relevance and Zarmik Moqataderi and Joseph Geisberg for help with the bioinformatic analysis and useful discussions. This work was funded by the Swiss National Science Foundation (Project P2FRP3_178090, to R.D.) and by an NIH research grant (CA 107486, to K.S.).
Footnotes
The authors declare no competing interests.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2014165117/-/DCSupplemental.
Data Availability.
Sequencing data for the transcriptional profiling have been deposited in the National Cancer for Biotechnology Information’s Gene Expression Omnibus database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE149141). All other study data are included in the main text and supporting information.
References
- 1.St. John T. P., Davis R. W., Isolation of galactose-inducible DNA sequences from Saccharomyces cerevisiae by differential plaque filter hybridization. Cell 16, 443–452 (1979). [DOI] [PubMed] [Google Scholar]
- 2.Schena M., Shalon D., Davis R. W., Brown P. O., Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270, 467–470 (1995). [DOI] [PubMed] [Google Scholar]
- 3.Lipshutz R. J., Fodor S. P., Gingeras T. R., Lockhart D. J., High-density synthetic oligonucleotide arrays. Nat. Genet. 21, 20–24 (1999). [DOI] [PubMed] [Google Scholar]
- 4.Wang Z., Gerstein M., Snyder M., RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 10, 57–63 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tirosh I., Wong K.-H., Barkai N., Struhl K., Extensive divergence of the yeast stress response through transitions between induced and constitutive activation. Proc. Natl. Acad. Sci. U.S.A. 108, 16693–16698 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hartwell L. H., Macromolecule synthesis in temperature-sensitive mutants in yeast. J. Bacteriol. 93, 1662–1670 (1967). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nusslein-Volhard C., Wieschaus E., Mutations affecting segment number and polarity in Drosophila. Nature 287, 795–801 (1980). [DOI] [PubMed] [Google Scholar]
- 8.Mohr S. E., Smith J. A., Shamu C. E., Neumuller R. A., Perrimon N., RNAi screening comes of age: Improved techniques and complementary approaches. Nat. Rev. Mol. Cell Biol. 15, 591–600 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karczewski K. J., Snyder M., Integrative omics for health and disease. Nat. Rev. Genet. 19, 299–310 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hirsch H. A., et al. , A transcriptional signature and common gene networks link cancer with lipid metabolism and diverse human diseases. Cancer Cell 17, 348–361 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou X., Liu J., Inferring gene dependency network specific to phenotypic alteration based on gene expression data and clinical information of breast cancer. PLoS One 9, e92023 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gupta S., et al. , Temporal expression profiling identifies pathways mediating effect of causal variant on phenotype. PLoS Genet. 11, e1005195 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feiglin A., Allen B. K., Kohane I. S., Kong S. W., Comprehensive analysis of tissue-wide gene expression and phenotype data reveals tissues affected in rare genetic disorders. Cell Syst. 5, 140–148.e2 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zarringhalam K., Degras D., Brockel C., Ziemek D., Robust phenotype prediction from gene expression data using differential shrinkage of co-regulated genes. Sci. Rep. 8, 1237 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nassiri I., McCall M. N., Systematic exploration of cell morphological phenotypes associated with a transcriptomic query. Nucleic Acids Res. 46, e116 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gallardo-Orihuela A., et al. , Transcriptional correlates of the pathological phenotype in a Huntington’s disease mouse model. Sci. Rep. 9, 18696 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iliopoulos D., Hirsch H. A., Struhl K., An epigenetic switch involving NF-kB, lin 28, let-7 microRNA, and IL6 links inflammation to cell transformation. Cell 139, 693–706 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iliopoulos D., Jaeger S. A., Hirsch H. A., Bulyk M. L., Struhl K., STAT3 activation of miR-21 and miR-181b, via PTEN and CYLD, are part of the epigenetic switch linking inflammation to cancer. Mol. Cell 39, 493–506 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ji Z., He L., Regev A., Struhl K., Inflammatory regulatory network mediated by the joint action of NF-kB, STAT3, and AP-1 factors is involved in many human cancers. Proc. Natl. Acad. Sci. U.S.A. 116, 9453–9462 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ji Z., et al. , Genome-scale identification of transcription factors that mediate an inflammatory network during breast cellular transformation. Nat. Commun. 9, 2068 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rotem A., et al. , Alternative to the soft-agar assay that permits high-throughput drug and genetic screens for cellular transformation. Proc. Natl. Acad. Sci. U.S.A. 112, 5708–5713 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soule H. D., et al. , Isolation and characterization of a spontaneously immortalized human breast epithelial cell line, MCF10. Cancer Res. 50, 6075–6086 (1990). [PubMed] [Google Scholar]
- 23.Jin Y., et al. , Mapping 3' mRNA isoforms on a genomic scale. Curr. Protoc. Mol. Biol. 110, 4.23.1–4.23.17 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data for the transcriptional profiling have been deposited in the National Cancer for Biotechnology Information’s Gene Expression Omnibus database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE149141). All other study data are included in the main text and supporting information.