SUMMARY
Neutralization of tumor necrosis factor (TNF) represents a widely used therapeutic strategy for autoimmune diseases including inflammatory bowel disease (IBD). However, the fact that many patients with IBD are non-responsive to anti-TNF therapies suggests the need for a better understanding of TNF signaling in IBD. Here, we show that co-deletion of TNF receptor 1 (TNFR1, Tnfrsf1a) in the Il10−/− spontaneous colitis model exacerbates disease, resulting in very-early-onset inflammation after weaning. The disease can be interrupted by treatment with antibiotics. The single deletion of TNFR1 induces subclinical colonic epithelial dysfunction and mucosal immune abnormalities, including accumulation of neutrophils and depletion of B cells. During the pre-disease period (before weaning), both Tnfr1−/− and Il10−/− Tnfr1−/− animals exhibit impaired expression of pro-inflammatory cytokines compared with wild-type and Il10−/− controls, respectively. Collectively, these results demonstrate the net anti-inflammatory functions of TNF/TNFR1 signaling through the regulation of colonic immune homeostasis in early life.
Graphical Abstract
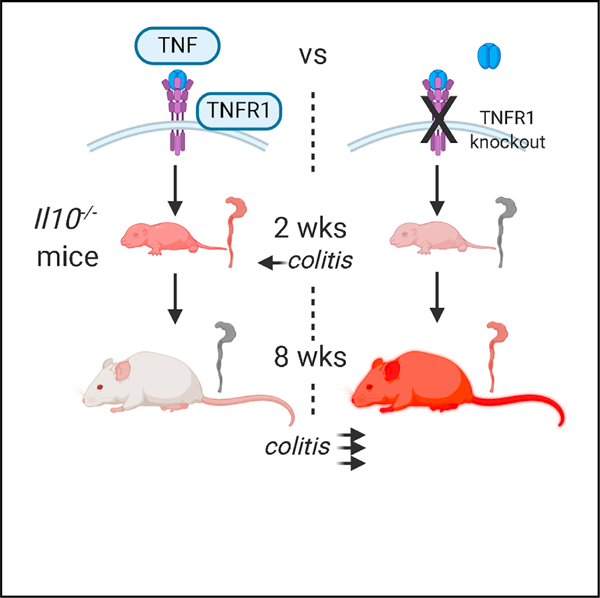
In Brief
Although anti-TNF therapies are used to treat colitis, Liu et al. demonstrate that colitis-susceptible mice deficient for TNF receptor 1 (TNFR1) paradoxically develop severe disease shortly after weaning. TNFR1 function can be traced back to its mediation of pro-inflammatory responses during a critical period of immune development in early life.
INTRODUCTION
The global incidence of inflammatory bowel disease (IBD) has increased steadily, especially in children under the age of 10 years (Benchimol et al., 2014; Kappelman et al., 2013; Schildkraut et al., 2013). A combination of host genetics, environmental exposures, intestinal mucosal dysfunction, and microbial dysbiosis contributes to IBD pathogenesis (Jostins et al., 2012). Although diverse genes and pathways regulate IBD susceptibility, their roles in immune development in early life and the biological mechanisms linking them to intestinal inflammation remain to be elucidated.
Tumor necrosis factor (TNF, formerly known as TNF-α) is a major therapeutic target in IBD. Pediatric patients with IBD have elevated circulating and intestinal levels of TNF (Breese et al., 1994; Murch et al., 1991). Recent genome-wide association studies (GWAS) have identified mutations in the TNF signaling pathway to confer risk for both ulcerative colitis and Crohn’s disease (Bank et al., 2014; Ferguson et al., 2009; Lappalainen et al., 2008; Li et al., 2016; McGovern et al., 2015; Pierik et al., 2004; Sashio et al., 2002; Waschke et al., 2005). The immediate therapeutic benefits of anti-TNF agents have suggested that TNF is a pro-inflammatory cytokine responsible for mucosal damage in IBD. However, the therapeutic effects of anti-TNF agents are short lived for many (Ben-Horin et al., 2014; Colombel et al., 2007; Ford et al., 2011; Schreiber et al., 2007). In mice, deletion of TNF or either of its transmembrane receptors, TNFR1 (p55) and TNFR2 (p75) (encoded by the Tnfrsf1a and Tnfrsf1b genes, respectively), has differing effects on disease susceptibility in established colitis models (Dubé et al., 2015; Ebach et al., 2005; Mizoguchi et al., 2008; Wang et al., 2012; Wang et al., 2013). Strikingly, loss of TNF-signaling-pathway members Tnf (Hale and Greer, 2012) or Tnfr2 (Punit et al., 2015) results in severe disease in the Il10−/− spontaneous colitis model (Kühn et al., 1993). By ablating a critical tolerogenic signal, interleukin 10 (IL-10), the Il10−/− model replicates key features of Crohn’s colitis, including the developmental dynamics of disease. However, the role of TNFR1 in the susceptibility to Il10−/− colitis has not, to our knowledge, been elucidated. It is not known whether TNFR1, on balance, would mediate the beneficial or deleterious effects of TNF signaling.
Here, we report severe, very-early-onset (VEO) colitis in Il10−/− Tnfr1−/− mice. Paradoxically, the early disease onset was preceded by impaired expression of pro-inflammatory cytokines during the period of life before weaning. TNFR1-mediated signaling in early life is, therefore, essential for the acquisition of mucosal tolerance.
RESULTS
Very-Early-Onset Colitis in Il10−/− Tnfr1−/− Mice
To characterize the role of TNFR1 in an animal model of spontaneous colitis, we bred Tnfr1−/− mice (Pfeffer et al., 1993) to Il10−/− mice (Kühn et al., 1993) to generate double-knockout animals. Consistent with previous reports (Bristol et al., 2000; Farmer et al., 2001; Mähler et al., 2002), Bl/6 Il10−/− mice are relatively resistant to colitis. Il10−/− Tnfr1−/− mice, however, developed severe, spontaneous, early-onset colitis, with a mixed mucosal infiltrate, cryptitis, abscesses, and epithelial hyperplasia. Although Il10−/− Tnfr1−/− animals at 2 (n = 8; Figures 1A and 1B) and 3 (n = 4, not shown) weeks old were spared colitis, whereas, at 4 weeks old, 8/13 (62%) of the Il10−/− Tnfr1−/− mice had histologic evidence of colitis (Figures 1C and 1D) (median histologic disease score: 7/15). At 6 weeks old, 4/7 (57%) of the Il10−/− Tnfr1−/− mice were colitic, and by 8 weeks old, 11/12 (92%) of the Il10−/− Tnfr1−/− mice had colitis (median histologic disease score: 8/15). Littermate Il10−/− mice had minimal disease (Figure 1E); moreover, heterozygous Il10−/− Tnfr1+/− littermates were spared severe disease and early mortality. Only 50% of Il10−/− Tnfr1−/− mice survived to 12 weeks old (Figures 1F, S1A, and S1B); their colons demonstrated increased RNA expression of cytokines, including Tnf (Figure 1G, from the NanoString assay) and increased crypt cell proliferation (phosphorylated histone H3 [pH-H3+] cells; Figure 1H) and apoptosis (TUNEL+ cells; Figure 1I). The disease affected the entire colon and cecum, resulting in serrated adenocarcinoma (Figure S1C), and retarded growth in both males and females (Figure S1D). This disease is, therefore, different from colitis in Il10−/− Tnfr2−/− mice, in which the cecum was not involved (Punit et al., 2015).
Figure 1. Il10−/− Tnfr1−/− Mice Develop Early-Onset, Severe Colitis at 4 Weeks Old.
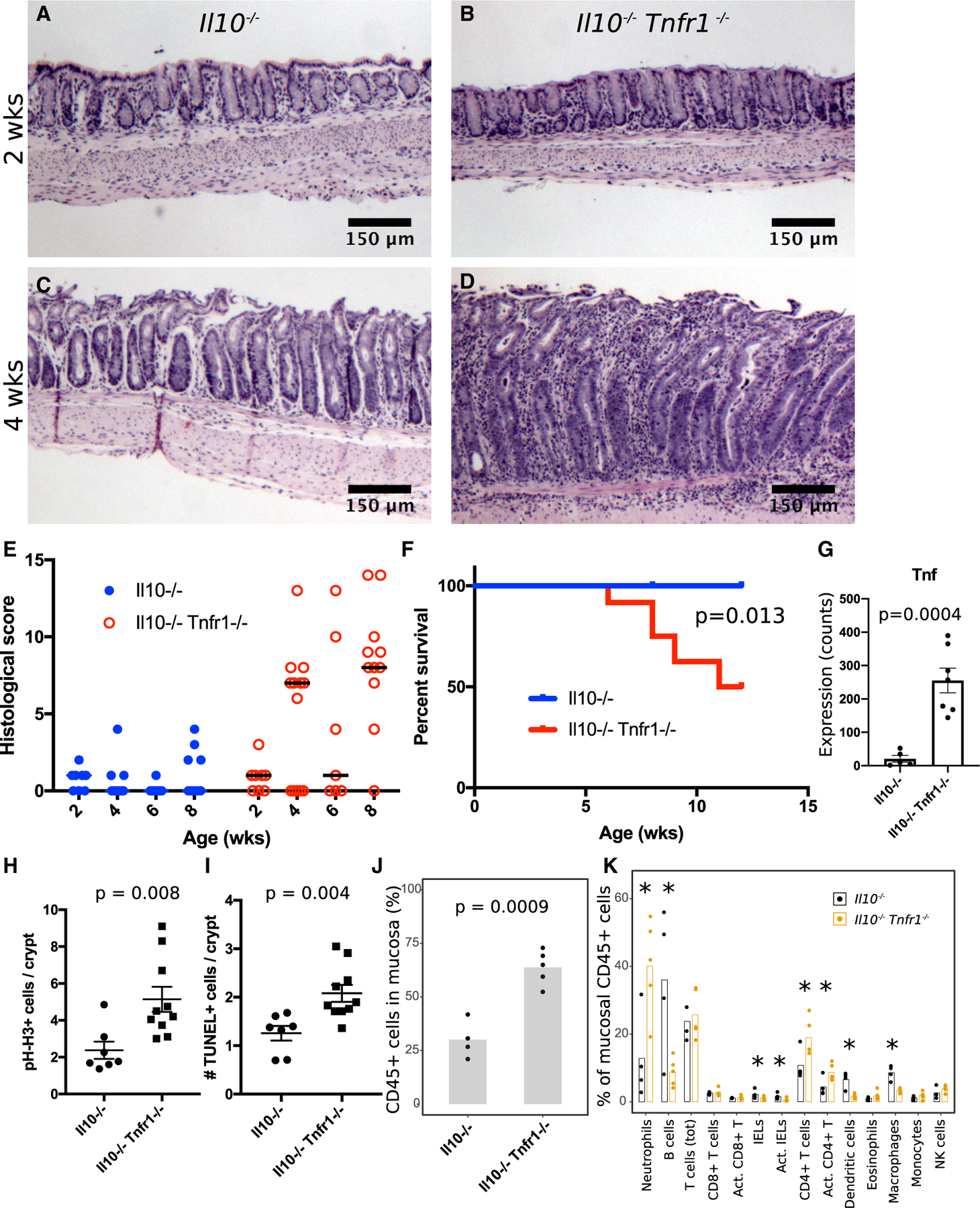
(A and B) Hematoxylin-and-eosin (H&E)-stained sections of pre-weaning 2-wk-old Il10−/− (A) (n = 7) and Il10−/− Tnfr1−/− (B) (n = 8) mice show normal distal colon.
(C and D) H&E sections of 4-week-old post-weaning mice reveal normal colon in the Il10−/− genotype (C) (n = 6) but colitis in the Il10−/− Tnfr1−/− genotype (D) (n = 13).
(E) Histological scoring of colitis severity (worst disease score is 15). Horizontal lines indicate median.
(F) Kaplan-Meier survival curve indicates early mortality in Il10−/− Tnfr1−/− mice.
(G) Nanostring analysis demonstrates upregulation of Tnf expression in Il10−/− Tnfr1−/− animals.
(H and I) Crypts in 12-wk-old Il10−/− Tnfr1−/− mice exhibited increased numbers of proliferating (pH-H3+) (H) and apoptotic (TUNEL+) (I) cells.
(J and K) Flow cytometric analysis of colonic mucosal scrapings demonstrates increased colonic mucosal infiltration of CD45+ hematopoietic cells (J) with increased immune cell subsets (K) in 8-wk-old Il10−/− Tnfr1−/− mice.
Scale bars: (A)–(D) 100 μm. *p < 0.05. See also Figures S1, S2, and S3. Error bars: standard error.
Because TNF signaling has integral roles in immune cell activation and specialization, we hypothesized that the loss of TNFR1 altered the relative proportions of the immune cells in colitis. We analyzed distal colonic mucosal scrapings by flow cytometry with a panel of 13 immune-cell-targeted antibodies (Figure S2) (Yu et al., 2016). Adult (8-week-old) Il10−/− Tnfr1−/− specimens showed elevated numbers of CD45+ (hematopoietic origin) cells compared with age- and litter-matched Il10−/− mice (Figure 1J), suggestive of increased immune cell infiltration into the mucosa. Within the CD45+ cell population, the relative proportions of neutrophils and activated CD4+ T cells were increased compared with those of the controls. However, several cell types showed a proportional reduction in the Il10−/− Tnfr1−/−animals: B cells, intraepithelial lymphocytes, dendritic cells, and macrophages (Figure 1K). Thus, the loss of TNFR1 results in a fundamental imbalance of immune cell subtypes in the context of inflammation.
To determine whether TNFR1 expression protected from colitis in a general setting of IL-10 signaling inhibition, we assessed the susceptibility of Tnfr1−/− mice to colitis induced by repetitive administration of anti-IL-10 receptor-targeted antibodies (Figure S3A) (Carvalho et al., 2012; Kullberg et al., 2006; Singh et al., 2016). Antibody-injected Tnfr1−/− (knockout) mice exhibited crypt loss and mucosal immune infiltration, hallmarks of colitis, whereas antibody-injected Tnfr1+/− (heterozygous) mice were relatively protected from colitis (Figure S3B), as assessed histologically (Figure S3C). Thus, TNFR1 restricts colitis associated with the loss of IL-10 signaling.
Antibiotics Induce Remission of Colitis
Because IBD is thought to represent an abnormal immune reaction against gastrointestinal commensals, we tested whether antibiotics would alter the severe colonic injury observed in Il10−/− Tnfr1−/− mice. We treated 8-week-old Il10−/− Tnfr1−/− mice with neomycin and metronidazole for 2 weeks. Distal colonoscopy performed before and immediately after treatment showed resolution of the disease in the antibiotic-treated group (Figure 2A) compared with the vehicle-treated controls (p = 0.0043; Figure 2B). The difference in outcomes was apparent histologically (p = 0.0042; Figures 2C and 2D) and was associated with reduced numbers of pH-H3+ (proliferative) and cleaved caspase-3+ (apoptotic) cells (Figure 2E). In a mixed cohort of adult Il10−/− (n = 7) and Il10−/− Tnfr1−/− (n = 2) mice, we found that antibiotic treatment reduced fecal lipocalin, a marker of neutrophil activity, to baseline uninflamed levels (<50 pg/mg stool) within the first week of treatment (Figure 2F). Lipocalin levels rebounded within 1 week after antibiotic withdrawal. We noted diarrhea in Il10−/− Tnfr1−/− mice, which precluded the comparison of absolute lipocalin levels between genotypes. Taken together, these results indicate that disease continuation in Il10−/− Tnfr1−/− mice requires neomycin/metronidazole-sensitive commensals and that colonic epithelial and neutrophil abnormalities in these mice result from host-microbe interactions.
Figure 2. Broad-Spectrum Antibiotic Treatment Improves Colitis in Il10−/− Tnfr1−/− Mice.
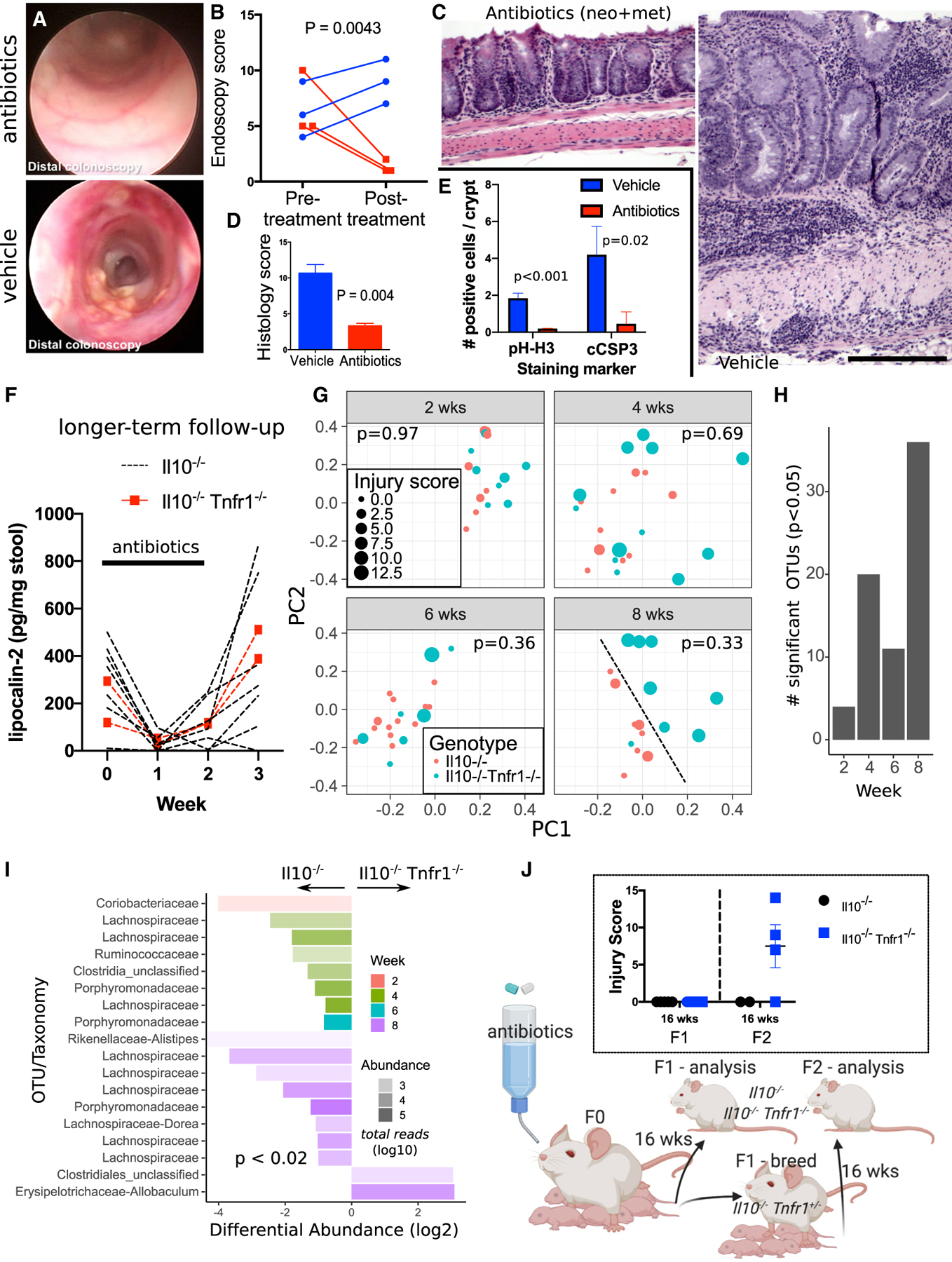
(A–E) Improved endoscopic appearance (A), endoscopic score (B), histology (C and D), and crypt cell dynamics (E) were observed in 8-week-old antibiotic-treated Il10−/− Tnfr1−/− mice (n = 3) compared with controls (n = 3).
(F) Fecal lipocalin levels diminished to nearly undetectable levels during antibiotic treatment of Il10−/− (n = 7) and Il10−/− Tnfr1−/− (n = 2) mice but quickly rebounded.
(G) 16S analysis of Il10−/− Tnfr1−/− and Il10−/− cecal microbiomes collected from 2-, 4-, 6-, and 8-week-old Il10−/− Tnfr1−/− and Il10−/− mice. Shown are weighted UniFrac distances plotted using multidimensional scaling. Each sample is coded by age (gray facets), genotype (color), and colitis score (size). At 8 weeks old, the genotypes could be qualitatively discriminated (dotted black line). The displayed p value for each age is computed by permuted analysis of variance using littermates, colitis severity, sex, and genotype conditioned on dam identity as model factors.
(H) Bar graph displays the total number of significant operational taxonomic units (OTUs) that are differentially abundant in Il10−/− versus Il10−/− Tnfr1−/− animals by age.
(I) OTUs with taxonomic classifications plotted to indicate their relative abundance in Il10−/− Tnfr1−/− versus Il10−/− samples. The transparency of the bars denotes the total abundance of the OTUs on a log scale (more transparent is less abundant).
(J) Depiction of the multigenerational experiment analyzing the effects of maternal antibiotic treatment on colitis development, with associated colitis scoring shown in the inset.
Scale bars: 200 μm. Error bars: standard error.
We next assessed whether the very-early-onset colitis observed in Il10−/− Tnfr1−/− mice was due to a distinct microbial ecosystem in early life. Using 16S sequencing, we profiled the luminal bacterial composition from the previously presented (Figure 1) cohort of littermate Il10−/− and Il10−/− Tnfr1−/− mice at different ages. Analysis of variance (permutational multivariate analysis of variance [PERMANOVA]) showed that microbial composition was significantly associated with age (p = 0.001) and dam (p = 0.01) but not genotype (p = 0.22) (Figure 2G). The abundances of few operational taxonomic units (OTUs) were significantly altered at 2 weeks old, but alterations increased after onset of colitis at 4 weeks old (Figure 2H). We note that at 2 weeks old, low-abundance Coriobacteriaceae were depleted in Il10−/− Tnfr1−/− samples (Figure 2I), consistent with this taxon’s reduced representation in pediatric IBD (Maukonen et al., 2015). At 8 weeks old, a qualitative difference in microbiome composition was observed (Figure 2G), but that difference was likely secondary to colitis. Colitic mice were depleted of Lachnospiraceae, a known producer of butyrate (Figure 2I) (Geirnaert et al., 2017; Surana and Kasper, 2017; Vital et al., 2014). Thus, a specific commensal signature did not cause exacerbated disease.
We next tested whether treatment of mice with antibiotics during the perinatal period would affect the onset of disease. Pregnant Il10−/− Tnfr1+/− dams were exposed to neomycin and metronidazole in their final week of pregnancy and up to 7 d postpartum. After treatment, F1 pups of all genotypes (Il10−/−, Il10−/− Tnfr1+/−, and Il10−/− Tnfr1−/−) remained free of disease to at least 16 weeks old (Figure 2J). Heterozygous Il10−/− Tnfr1+/− pups were subsequently interbred, and their F2 pups of the Il10−/− Tnfr1−/− genotype exhibited colitis at 16 weeks old. Thus, the early onset of colitis in Il10−/− Tnfr1−/− mice can be interrupted by maternal treatment with antibiotics.
Colonic Mucosal Dysfunction in Tnfr1−/− Mice
To determine the specific contribution of TNFR1 to the colitic disease process, we analyzed the physiological effects of TNFR1 genetic loss in isolation, using Il10+/+ Tnfr1−/− mice (or simply Tnfr1−/− mice). The fecal lipocalin levels of Tnfr1−/− mice at 12 weeks old were all <50 pg/mg stool (n = 7), suggesting that these mice did not have overt colitis. However, we found crypt fission, dropout, and branching (Figure 3A) in foci in 3/7 (43%) Tnfr1−/− mice. Moreover, at 4, 8, and 16 weeks old, Tnfr1−/− mice had significantly increased serum recovery of enemaadministered fluorescein isothiocyanate (FITC)-dextran (Figure 3B). Tnfr1−/− colonic crypts at 4 and 12 weeks old exhibited modestly elevated numbers of proliferating pH-H3+ cells (Figure 3C), and crypt abnormalities could also be identified in 4-week-old mice (Figure 3D; in 4/8 mice). At 12 weeks old, Tnfr1−/− mice had significantly (p = 0.03) increased numbers of DNA-damaged phosphorylated histone 2A.X (pH2A.X)-positive epithelial cells and phosphorylated STAT3-positive epithelial cells (Figure 3E). In contrast to Il10−/− Tnfr1−/− mice, intra-crypt staining for cleaved caspase-3 apoptotic bodies was not observed in Tnfr1−/− mice (not shown). TNFR1, therefore, regulates colonic epithelial function (e.g., morphology, permeability, and DNA-damage signaling) and repair (e.g., proliferation and STAT signaling) from a young age.
Figure 3. Adult Tnfr1−/− Mice Exhibit Epithelial Dysfunction and Alterations in Colonic Mucosal Immune Cell Representation.
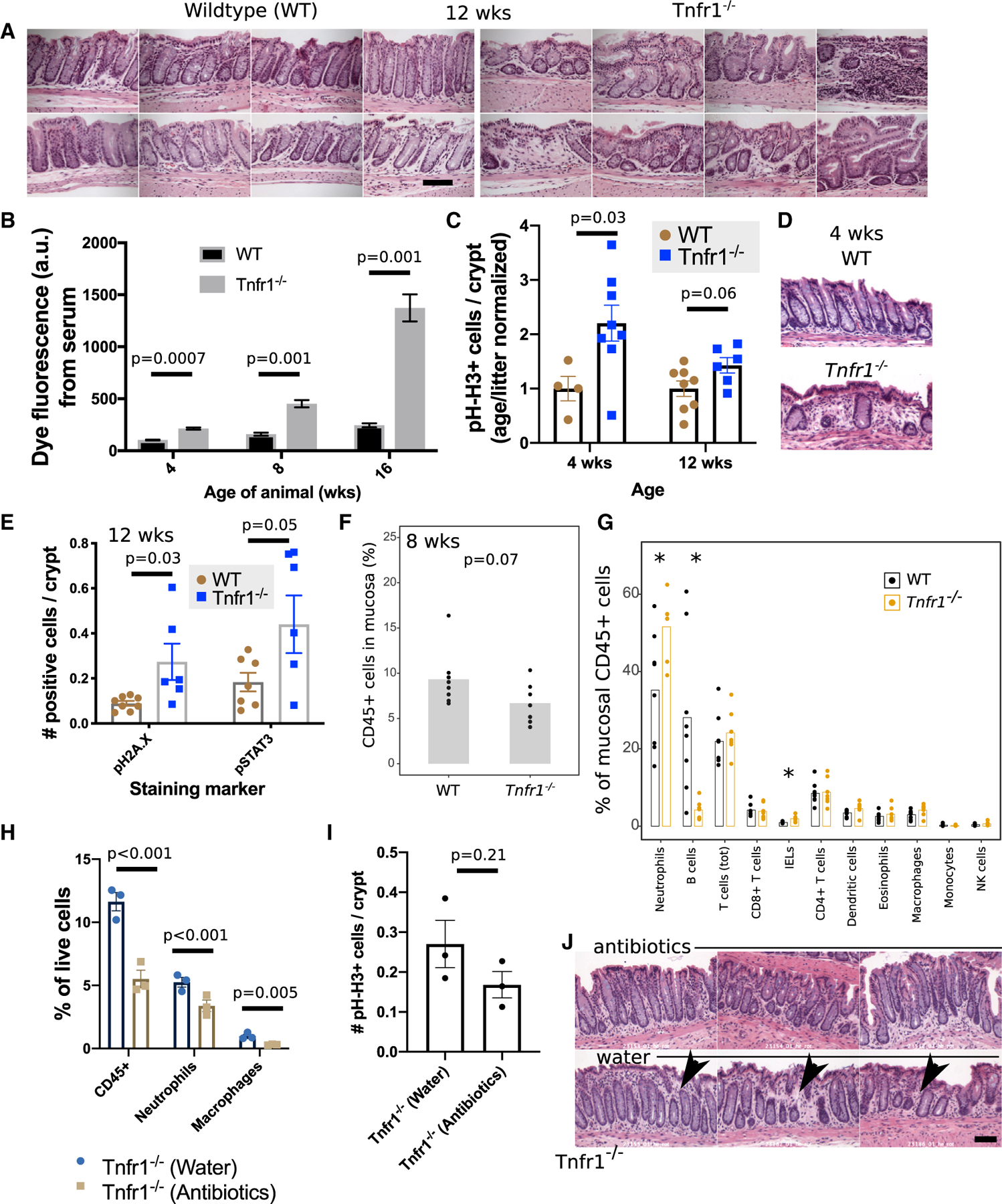
(A) Image montages show normal crypt structures in wild-type mice (n = 8) and focal abnormalities in 8-wk-old Tnfr1−/− mice (n = 7).
(B) Tnfr1−/− mice at 4, 8, and 12 weeks old exhibited increased colonic permeability at 30 min after rectal instillation of 4 kDa FITC-dextran. The graph is representative of three different experiments.
(C) Trending increase in pH-H3+ proliferative cells in knockout mice at 4 and 12 weeks old.
(D) H&E images of the colonic mucosa at 4 weeks old demonstrate foci of altered crypt patterning in Tnfr1−/− mice.
(E) Adult 12-week-old Tnfr1−/− colonic epithelium also had increased numbers of pH2A.X+ and pSTAT3+ cells.
(F and G) Flow cytometry reveals a trend toward decreased hematopoietic cell numbers (F) in the colonic mucosa of 8-week-old Tnfr1−/− mice. This was driven by increased neutrophils and a near-total loss of B cells (G).
(H) Flow cytometric assessment of antibiotic-treated Tnfr1−/− mice reveals reduced numbers of mucosal immune cells.
(I) Changes to pH-H3 staining after antibiotic treatment were more modest and insignificant.
(J) Qualitative reductions (arrowheads) in the density of inter-crypt cells were noted in H&E-stained sections of antibiotic-treated samples.
Scale bars: (A) 100 μm; (D) and (J) 50 μm, *p < 0.05. Error bars: standard error.
We also found significant immune defects through flow cytometry. There was a trending reduction (p = 0.07) in the total number of mucosal immune cells in 8-week-old Tnfr1−/− mice (Figure 3F). Although the proportions of T cells were similar between Tnfr1−/− and wild-type mice, the Tnfr1−/− mice showed an ~90% loss of B cells, consistent with prior reports of the TNFR1 role in the establishment of germinal centers (Le Hir et al., 1996; Matsumoto et al., 1996; Pasparakis et al., 1996). Knockout mice showed a higher percentage of neutrophils (Figure 3G). Treatment of adult Tnfr1−/− mice with neomycin and metronidazole antibiotics for 2 weeks reduced the total representation of CD45+ hematopoietic cells, neutrophils (CD45+ Ly6G+), and macrophages (CD45+ SSChi CD11b/c+ IA/IE+ CD24−) (Figure 3H). However, antibiotic-driven reductions in colonic epithelial pH-H3 staining did not reach significance (Figure 3I). A reduced inter-crypt cellularity was histologically apparent in antibiotic-treated animals (Figure 3J). Thus, live commensals contribute to the mucosal defects associated with the loss of TNFR1.
Loss of TNFR1 Reduces Early-Life Cytokine Expression
The very-early onset of disease in Il10−/− Tnfr1−/− mice occurs shortly after the completion of the weaning process. We ordinarily separate pups from the dam at 3 weeks of age. Delaying that separation by 1 week (from 3 weeks old to 4 weeks old) did not prevent disease onset at 4 weeks (data not shown). Thus, the cause of disease cannot be reduced to this single ‘‘trigger’’ of maternal separation.
We next asked whether the effects of TNFR1 loss could be discerned in the pre-disease, early-life period. By high-throughput expression profiling, we examined the full-thickness colons of wild-type, Tnfr1−/−, Il10−/−, and Il10−/− Tnfr1−/− mice at ages corresponding to the pre-colitic (2 weeks) and colitic (8 weeks) states. In RNA sequencing (RNA-seq) studies comparing Il10−/− Tnfr1−/− and Il10−/− mice at 2 weeks old, we observed a profound downregulation of classical inflammatory markers, including Nos2, Cxcl1, Saa1, and calprotectin (S100a8 and S100a9), in the double-knockout mice (Figure 4A). We analyzed those cytokines as a group, the ‘‘TNFR1-associated early-life immune module,’’ and calculated a score (−1.7 ± 0.35, their mean logarithmic expression ratio between TNFR1-deficient and TNFR1-expressing conditions), which quantified their overall change (Figure 4B). That score was significantly less than zero (p = 3 × 10−5), supporting the overall reduced expression of those cytokines in Il10−/− Tnfr1−/− mice at 2 weeks of age. At 8 weeks old, these immune markers had a positive (p = 0.003) expression score of 3.4 ± 1.0 in Il10−/− Tnfr1−/− (versus age-matched Il10−/−) animals, consistent with their broad upregulation in colitis. This pattern of changes was preserved in Tnfr1−/− mice that had genetically functional IL-10, although the magnitude of the changes was less. Adult Tnfr1−/− mice had elevated expression of neutrophil-associated genes, such as the calprotectin subunits (S100a8, S100a9) and Duox2. In contrast, pre-weaning Tnfr1−/− mice at 2 weeks old exhibited reduced transcription of Il1b (IL-1b) (Figure 4A). qPCR studies showed that adult heterozygous Tnfr1+/− mice had similar calprotectin expression as wild-type littermates (Figure S4A).
Figure 4. TNFR1-Deficient Animals Show Early-Life Defects in Colonic Immune Response.
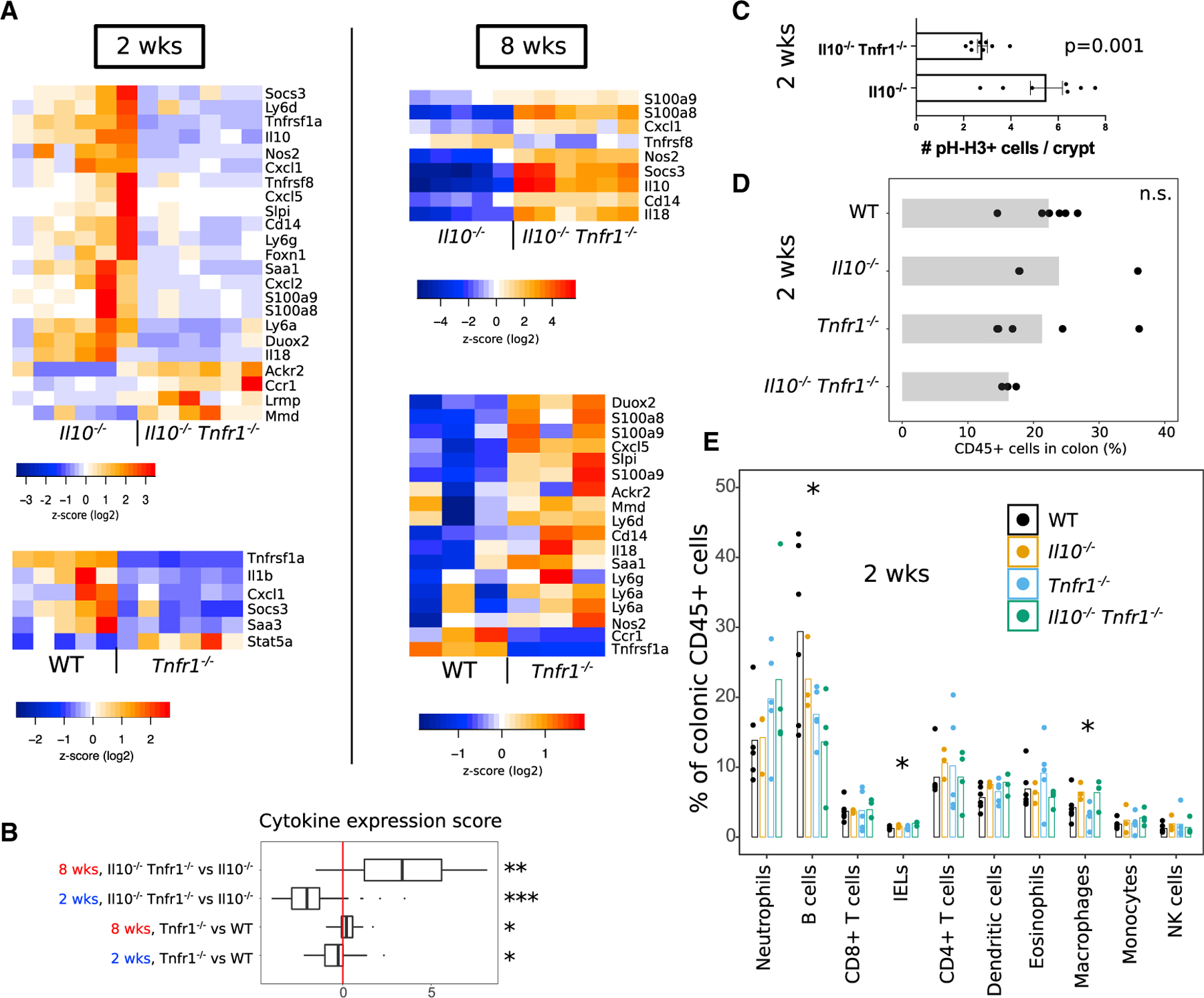
(A) Heatmaps summarize cytokine-expression levels obtained from RNA-seq/NanoString analysis of colons from wild-type (WT), Tnfr1−/−, Il10−/−, and Il10−/− Tnfr1−/− animals at 2 weeks (pre-disease) and 8 weeks (post-disease onset) old. Overall cytokine expression was decreased in TNFR1-deficient animals at 2 weeks of age but increased in those animals at 8 weeks old. Note that reads for Il10 were still detected in Il10−/− animals because of continued production of mRNA encoding of an early-termination codon.
(B) The boxplot shows a summarized score of cytokine expression changes across comparisons of genotypes and ages. The lower and upper hinges represent the 25th and 75th percentiles. Whiskers denote the range of the data, excepting outlying points beyond 150% of the interquartile range. All values were obtained from RNA-seq except for the comparison of Il10−/− Tnfr1−/− versus Il10−/− mice at 8 weeks old (NanoString). Significance of scores was tested using the t test against 0 (no change).
(C) Il10−/− Tnfr1−/− animals at 2 weeks old show reduced pH-H3+ cell counts per crypt.
(D and E) Flow cytometric analysis of whole colons collected from 2-week-old mice shows overall similarity of colonic hematopoietic (CD45+) cell counts (D) but loss of B cells in TNFR1-deficient animals (E).
Significance was evaluated using ANOVA. *p < 0.05. See also Figure S4. Error bars: standard error.
Cytokine changes were associated with functional defects. There was more epithelial pH-H3 (proliferative) staining in 2-week-old Il10−/− colons versus Il10−/− Tnfr1−/− colons (Figure 4C), in marked contrast to results in 8-week-old mice (Figure 1H). On flow-cytometric analysis, we found similar proportions of CD45+ cells (Figure 4D) and neutrophils (Figure 4E) in 2-week-old animals across all genotypes. However, reduction in B cell numbers in TNFR1-deficient animals was already evident at 2 weeks of age (Figure 4E). Thus, loss of TNFR1 results in profound immune and epithelial defects in early life.
We ruled out that transcriptional changes in Tnfr1−/− mice were compensated for by TNFR2. We compared transcriptomic profiles of adult Tnfr1−/− mice and Tnfr2−/− mice (Punit et al., 2015). As shown in Figure S4B, there was little overlap (4%) of differentially expressed transcripts in Tnfr1−/− and Tnfr2−/− colons. Among the few co-regulated transcripts, there was a significant negative correlation in their direction of regulation (Figure S4C). The loss of Tnfr1 did not affect expression of Tnfr2 and vice versa. Notably, Saa2 and Duox2, associated with abnormal mucosal immune responses to microbiota (Eckhardt et al., 2010; Grasberger et al., 2015), were both upregulated in the absence of either TNFR1 or TNFR2.
DISCUSSION
Here, we show that the genetic loss of TNFR1 results in increased susceptibility to colitis. The early age of disease onset in Il10−/− Tnfr1−/− mice, after weaning, may be functionally equivalent to infantile or toddler colitis in humans (assuming full weaning at 1–2 years of age). In rodents, at that age, the colonic mucosa undergoes histological maturation (Walthall et al., 2005). During the simultaneous immune ‘‘weaning reaction,’’ pro-inflammatory signals act against intestinal luminal contents to ultimately promote tolerance through the establishment of immunosuppressive cell populations (Al Nabhani et al., 2019; Olszak et al., 2012; Pié et al., 2004; Redhu et al., 2017; Scheer et al., 2017). The results of our study can be interpreted within that framework and demonstrate that TNFR1-mediated pro-inflammatory signaling, including upregulation of Il1b, in early life is essential for the weaning reaction (Pié et al., 2004). These results demonstrate how immunodeficiency during early life can predispose animals toward later autoimmunity, as hinted at by studies of other genetic immunodeficiencies (Glocker and Grimbacher, 2012; Mombaerts et al., 1993; Sadlack et al., 1993; Salzer et al., 2014; Tegtmeyer et al., 2017).
The exacerbation of colitis with TNFR1 loss in mice could partially model the uneven clinical efficacy of anti-TNF antibodies in IBD (Gratz et al., 2002; Kullberg et al., 2001; Scheinin et al., 2003), increased risk for human IBD associated with genetic perturbation of TNF receptors, and new-onset IBD in autoimmune patients treated with anti-TNF (Korzenik et al., 2019; Toussirot et al., 2012; Üsküdar Cansu et al., 2019). Relevant to this study, very-early-onset IBD is frequently linked to the loss of the IL-10 receptor (Bianco et al., 2015) (Begue et al., 2011; Beser et al., 2015; Kotlarz et al., 2012; Moran et al., 2013; Pigneur et al., 2013). Elucidating key functions in TNF receptor signaling in intestinal development and inflammation may have therapeutic benefits.
Limitations of Study
First, it is premature to generalize our findings directly to human anti-TNF therapies, which may work through distinct mechanisms (e.g., Atreya et al., 2011). Second, we have had only limited success in pharmacological manipulation of the weaning reaction in colitis. Third, we do not know whether immune cell alterations and epithelial dysfunction in Tnfr1−/− animals are caused by one or the other. Future studies will need to elaborate on these aspects of TNF signaling in colitis.
STAR★METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Brent Polk (dbpolk@chla.usc.edu).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
The RNA-seq datasets generated during this study are available at NCBI Gene Expression Omnibus (accession numbers): GSE107933, GSE155654, GSE155626).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
Mice were maintained humanely, in accordance with the rules and regulations of the Institutional Animal Care and Use Committee (IACUC) of Children’s Hospital Los Angeles (CHLA). This study was approved by the CHLA IACUC under the internal protocol number 288. Mice were anesthetized with isoflurane and euthanized via cervical dislocation prior to dissection.
Wild-type (WT), Il10−/− (stock #002251), and Tnfr1−/−(stock #002818) mice on the C57BL/6J background were obtained from Jackson Laboratory. Il10−/− Tnfr1−/− mice and control Il10−/− littermates were generated from Il10−/− Tnfr1+/− parents. Experimental and control groups of mice were co-housed for > 4 weeks prior to experimentation, and co-housing was maintained during experimentation. Both male and female mice were included in the study. Details regarding the ages of mice used in experiments are given in the description of Results.
METHOD DETAILS
Tissue collection
Colons and ceca were carefully freed from the abdominal cavity of euthanized mice, opened longitudinally, and cleaned of fecal contents. Luminal contents of the ceca were collected for bacterial 16S rRNA sequencing. Colons were opened and splayed; a thin longitudinal strip representing the full proximal-distal length of each colon was removed and preserved for RNA isolation. The remaining tissue was fixed overnight in 10% neutral buffered formalin at room temperature. For histological assessment, colons were washed with 50% ethanol, dehydrated in an ascending ethanol series, cleared with xylenes, and embedded in molten paraffin. Thin (5 μm) sections were prepared on a microtome and stored until their usage in staining procedures.
Histochemistry
Blinded to sample origin, assessment of colitis severity was made on hematoxylin and eosin-stained sections of murine colon by MKW using previously validated and published methodologies (Dubé et al., 2012; Zhang et al., 2012). Histology was scored based on 5 categories: enterocyte loss, crypt inflammation, lamina propria mononuclear cells, polymorphonuclear cells, and hyperplasia. Each category is awarded a score from 0 to 3, with higher score indicating more severe disease. A maximum score of 15 is assigned. For quantification of epithelial cell markers, label-positive cells in 30–100 crypts adjacent to the anal verge were counted and divided by the total number of counted crypts to yield a per-crypt number.
Immunohistochemistry was performed using standard protocols. Primary antibodies were incubated at 4○C overnight. Primary antibodies used were rabbit anti-pH2A.X (Ser139) (Cell Signaling Technology #9718S, 1:2,000 dilution), rabbit anti-pHH3 (Ser10) (Cell Signaling Technology #9701S, 1:500 dilution), rabbit anti-pSTAT3 (Tyr705) (Cell Signaling Technology #9145, 1:100 dilution), and rabbit anti-cCSP3 (Cell Signaling Technology #9661S, 1:200 dilution).
Flow Cytometry
Colons were freshly dissected from mice. If the colon was isolated from an adult (8-wk-old) animal, the mucosal layer was separated from the muscle layer using fine forceps. The mucosal piece was minced into 2-mm2 pieces. If the colon was isolated from an unweaned (2-wk-old) mouse, the full-thickness organ was immediately minced into 2-mm2 pieces.
Tissue was incubated with prewarmed digestion solution (0.2 Wunsch units/ml Liberase TM + 200 Kuntz units/ml DNase I in DMEM/F12 + 15 mM HEPES) for 30 min at 37○C with continuous agitation at 180 rpm. After trituration of the tissue and its passage through a 70-μm-pore cell strainer, the tissue was washed with DMEM:F12 supplemented with 10% FBS, and then also with HEPES-buffered saline (Liu et al., 2012) supplemented with 0.5% BSA. All subsequent washes and stainings were performed in HBS+0.5% BSA. The tissue was stained with 0.1 μM DAPI, blocked for 15 min at 4○C with a solution composed of 5% mouse/rat serum supplemented with mouse Fc block (anti-CD16/32 antibody, Biolegend ‘‘truStain fx’’), and probed with a mixture of preconjugated antibodies for 30 min at 4○C. Antibodies (working dilutions) were targeted to CD45 (1:250), Ly6G (1:200), CD11b (1:200), CD11c (1:100), IA/IE (1:1,000), CD64 (1:400), CD24 (1:500), Ly6C (1:200), CD69 (1:200), CD8 (1:100), CD4 (1:200), TCRd (1:200), and CD3 (1:200). After washes, cells were analyzed on a BD LSR II. Compensation was adjusted using references obtained by the analysis of antibodies bound to Ultracomp eBeads (ThermoFisher Scientific).
Analysis of flow cytometric data was performed using FlowJo. Gates were adjusted for each experimental day but were consistent between experimental and control samples. Data were pooled across 3–4 experimental days per genotype/age comparison.
Collection and analysis of stool
Stool pellets were collected from mice by manual restraint which typically induced defecation within 30 s. A clean Eppendorf tube was held beneath the anal opening. Pellets were frozen at −80○C and analyzed for fecal lipocalin-2 levels using a commercial ELISA kit and according to the manufacturer’s instructions (R&D Systems, cat# DY1857).
Antibiotic treatment
Il10−/− Tnfr1−/− mice at 8 weeks of age were assigned to either ‘Vehicle’ or ‘Antibiotics’ treatment group (n = 3 in each group). Mice received either water (‘Vehicle’) or broad-spectrum antibiotics for 2 weeks (Neomycin sulfate 500mg tablets purchased from Hi-Tech Pharmacal, Metronidazole 500mg/100ml purchased from Claris Lifescience) ad libitum. Both antibiotics were mixed with drinking water at 1 g/L. Distal colonoscopy on deeply anesthetized mice was performed pre- and post-treatment using a flexible instrument (Karl Storz). Mice were euthanized post-treatment at 10 weeks of age, and colon histology was scored as previously described (Dubé et al., 2012; Punit et al., 2015) to determine colitis severity.
16S sequencing
16S rRNA libraries were prepared from flash-frozen cecal contents. Cecal content was collected from co-housed littermates of Il10−/− and Il10−/− Tnfr1−/− mice euthanized at 2, 4, 6, and 8 weeks. Cecal content was frozen immediately after collection and stored at −80○C. Cecal microbiome composition was determined by sequencing the 16S ribosomal RNA gene. Sequencing was performed on the Illumina MiSeq platform (Diversity assay 2x300bp 20K bTEFAP® at MR DNA lab (Shallowater, TX USA)).
Expression analysis of immune-related transcripts
RNA was extracted from colon tissues using standard RNA extraction techniques (PureLink® RNA Mini Kit from Ambion) and assayed for mRNA expression of targeted immune genes using the Nanostring technology (nCounter Mouse Immunology Panel, 561 genes).
RNA-Seq
mRNA transcripts were purified from total colonic RNA using oligo-dT coated beads, sheared, and prepared for 2x75 bp sequencing on a HiSeq 4000 (Illumina). Library preparation and sequencing were performed by SeqMatic, LLC (Fremont, CA USA).
Barrier function assay
WT and Tnfr1−/− mice were co-housed for minimum of 1-week prior to experiment. Mice were sedated and a 0.2 mL volume of fluorescein (FITC)-dextran (MW 3000, Invitrogen, 50 mg/ml solution in PBS) was instilled into the rectum with a 3.5 Fr catheter. Blood was obtained 30 minutes later through a retro-orbital blood draw. Plasma fluorescence was measured in a plate reader (excitation 490nm; emission 520nm).
Anti-IL10R model of colitis
Co-housed Tnfr1+/− and Tnfr1−/− mice were intraperitoneally injected, on alternating sides of body, with 1 mg/mouse/dose of anti-IL10R antibody once per week, for a total of 3 injections. Injections were begun when mice were 3 wks old. Euthanasia and postmortem analyses were performed at 6 wks of age.
QUANTIFICATION AND STATISTICAL ANALYSIS
Gene abundance estimates
Transcript quantification and significance testing from RNA-Seq data were performed using the paired-end read option on kallisto/sleuth (Bray et al., 2016). Differential expression of expression data obtained using the Nanostring platform was computed using nSolver version 3.0 software (Nanostring).
Operational taxonomic units (OTUs) were generated from 16S sequences by clustering at 3% divergence (97% similarity) in mothur (Schloss et al., 2009). Final OTUs were taxonomically classified against a curated database derived from the Ribosomal Database Project (Cole et al., 2014) and NCBI. Additional analyses were performed using the ‘phyloseq’ (v1.19.1) (McMurdie and Holmes, 2013) and ‘vegan’ (v2.4–2) (Dixon, 2003) packages in R (version 3.3). Random forest regression was used to predict colitis score with OTU relative abundances as covariates (‘randomForest’ package v4.6). Only OTUs with > = 1% relative abundance in at least 5 samples were used. Linear discriminant analysis was performed using LEfSe (Segata et al., 2011).
Statistical tests
Data are presented as individual points with summary statistics reporting the mean and standard error of the mean. Significance was evaluated using the t test, unless otherwise noted.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| FITC rat anti-CD45 | Biolegend | 103107; RRID: AB_312972 |
| Alexa Fluor 700 rat anti-Ly6G | BD Biosciences | 561236; RRID: AB_10611860 |
| APC-Cy7 rat anti-CD11b | BD Biosciences | 561039; RRID: AB_2033993 |
| APC hamster anti-CD11c | BD Biosciences | 561119; RRID: AB_10562405 |
| PE rat anti-IA/IE | BD Biosciences | 562010; RRID: AB_10893194 |
| PE-Cy7 mouse anti-CD64 | Biolegend | 139313; RRID: AB_2563903 |
| BV421 rat anti-CD24 | BD Biosciences | 562563; RRID: AB_2737002 |
| PerCP-Cy5.5 rat anti-Ly6C | ThermoFisher Scientific | 45-5932-80; RRID: AB_2723342 |
| BV786 hamster anti-CD69 | BD Biosciences | 564683; RRID: AB_2738890 |
| BV510 rat anti-CD8a | Biolegend | 100751; RRID: AB_2561389 |
| BV605 rat anti-CD4 | Biolegend | 100451; RRID: AB_2564591 |
| PE-CF594 hamster anti-gdTCR | BD Biosciences | 563532; RRID: AB_2661844 |
| BUV395 hamster anti-CD3ε | BD Biosciences | 563565; RRID: AB_2738278 |
| Rabbit anti-pH2A.X (Ser139) | Cell Signaling Technology | 9718S; RRID: AB_2118009 |
| Rabbit anti-pH-H3 (Ser10) | Cell Signaling Technology | 9701S; RRID: AB_331535 |
| Rabbit anti-pSTAT3 (Tyr705) | Cell Signaling Technology | 9145; RRID: AB_2491009 |
| Rabbit anti-cleaved CSP3 | Cell Signaling Technology | 9661S; RRID: AB_2341188 |
| truStain fcX (rat anti-CD16/32) | Biolegend | 101320; RRID: AB_1574975 |
| Rat anti-mouse IL-10R (CD210), 1B1.3A | Bio X Cell | BE0050; RRID: AB_1107611 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Neomycin sulfate – 500 mg tablets | Hi-Tech Pharmacal | 50383-565-10 |
| Metronidazole – 500 mg/100 mL | Claris Lifesciences | 36000-001-24 |
| 4 kDa FITC-dextran | ThermoFisher Scientific | D3305 |
| Liberase TM | Sigma-Aldrich | 5401119001 |
| DNase I | Sigma-Aldrich | D5025 |
| Ultracomp eBeads | ThermoFisher Scientific | 01-2222-41 |
| Critical Commercial Assays | ||
| 16S library preparation | MR DNA (Molecular Research) | bTEFAP |
| mRNA-Seq library preparation (Illumina) | SeqMatic | 20020594 |
| nCounter Mouse Immunology Panel | Nanostring | XT-CSO-MIM1-12 |
| Mouse Lipocalin-2/NGAL DuoSet ELISA | R&D Systems | DY1857 |
| Deposited Data | ||
| RNA-Seq (Tnfr1−/− versus WT, 8 wks of age) | Gene Expression Omnibus (GEO) | GSE107933 |
| RNA-Seq (Tnfr1−/− versus WT, 2 wks of age) | Gene Expression Omnibus (GEO) | GSE155654 |
| RNA-Seq (Il10−/− Tnfr1−/− versus Il10−/−, 2 wks of age) | Gene Expression Omnibus (GEO) | GSE155626 |
| Experimental Models: Organisms/Strains | ||
| Mice: C57Bl6/J | Jackson Labs | 000664 |
| Mice: Il10−/− | Jackson Labs | 002251 |
| Mice: Tnfr1−/− | Jackson Labs | 002818 |
| Software and Algorithms | ||
| Kallisto/sleuth v0.44.0 | Pachter Lab | https://pachterlab.github.io/kallisto/ |
| nSolver v3.0 | Nanostring | https://www.nanostring.com/products/analysis-software/nsolver |
| mothur v1.40.1 | Schloss Lab | https://mothur.org/ |
| FlowJo v10.4 | FlowJo | https://www.flowjo.com/ |
Highlights.
Il10−/− Tnfr1−/− mice exhibit severe colitis beginning shortly after weaning
Colitis is dependent on the microbiome but is not taxa specific
Tnfr1−/− mice exhibit colonic immune dysregulation and abnormal epithelium
Tnfr1−/− mice have reduced cytokine expression during a critical weaning period
ACKNOWLEDGMENTS
This study was supported by the U.S. National Institutes of Health (R01-DK056008 and R01-DK108648, to D.B.P.), the Crohn’s and Colitis Foundation (career development award to C.Y.L., postdoctoral fellowship to P.E.D., and senior research award to D.B.P.), and the California Institute for Regenerative Medicine (postdoctoral fellowship to C.Y.L.). We would like to thank Tsen-Yin Lin, Michael Sheard, Elizabeth Lopez, Gricelda Vasquez, and Chris Escolano for technical advice and care of animals.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.108275.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Al Nabhani Z, Dulauroy S, Marques R, Cousu C, Al Bounny S, Dejardin F, Sparwasser T, Berard M, Cerf-Bensussan N, and Eberl G (2019). A weaning reaction to microbiota is required for resistance to immunopathologies in the adult. Immunity 50, 1276–1288.e1275. [DOI] [PubMed] [Google Scholar]
- Atreya R, Zimmer M, Bartsch B, Waldner MJ, Atreya I, Neumann H, Hildner K, Hoffman A, Kiesslich R, Rink AD, et al. (2011). Antibodies against tumor necrosis factor (TNF) induce T-cell apoptosis in patients with inflammatory bowel diseases via TNF receptor 2 and intestinal CD14+ macrophages. Gastroenterology 141, 2026–2038. [DOI] [PubMed] [Google Scholar]
- Bank S, Skytt Andersen P, Burisch J, Pedersen N, Roug S, Galsgaard J, Ydegaard Turino S, Brodersen JB, Rashid S, Kaiser Rasmussen B, et al. (2014). Polymorphisms in the inflammatory pathway genes TLR2, TLR4, TLR9, LY96, NFKBIA, NFKB1, TNFA, TNFRSF1A, IL6R, IL10, IL23R, PTPN22, and PPARG are associated with susceptibility of inflammatory bowel disease in a Danish cohort. PLoS ONE 9, e98815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begue B, Verdier J, Rieux-Laucat F, Goulet O, Morali A, Canioni D, Hugot JP, Daussy C, Verkarre V, Pigneur B, et al. (2011). Defective IL10 signaling defining a subgroup of patients with inflammatory bowel disease. Am. J. Gastroenterol 106, 1544–1555. [DOI] [PubMed] [Google Scholar]
- Ben-Horin S, Kopylov U, and Chowers Y (2014). Optimizing anti-TNF treatments in inflammatory bowel disease. Autoimmun. Rev 13, 24–30. [DOI] [PubMed] [Google Scholar]
- Benchimol EI, Manuel DG, Guttmann A, Nguyen GC, Mojaverian N, Quach P, and Mack DR (2014). Changing age demographics of inflammatory bowel disease in Ontario, Canada: a population-based cohort study of epidemiology trends. Inflamm. Bowel Dis 20, 1761–1769. [DOI] [PubMed] [Google Scholar]
- Beser OF, Conde CD, Serwas NK, Cokugras FC, Kutlu T, Boztug K, and Erkan T (2015). Clinical features of interleukin 10 receptor gene mutations in children with very early-onset inflammatory bowel disease. J. Pediatr. Gastroenterol. Nutr 60, 332–338. [DOI] [PubMed] [Google Scholar]
- Bianco AM, Girardelli M, and Tommasini A (2015). Genetics of inflammatory bowel disease from multifactorial to monogenic forms. World J. Gastroenterol 21, 12296–12310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray NL, Pimentel H, Melsted P, and Pachter L (2016). Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol 34, 525–527. [DOI] [PubMed] [Google Scholar]
- Breese EJ, Michie CA, Nicholls SW, Murch SH, Williams CB, Domizio P, Walker-Smith JA, and MacDonald TT (1994). Tumor necrosis factor alpha-producing cells in the intestinal mucosa of children with inflammatory bowel disease. Gastroenterology 106, 1455–1466. [DOI] [PubMed] [Google Scholar]
- Bristol IJ, Farmer MA, Cong Y, Zheng XX, Strom TB, Elson CO, Sundberg JP, and Leiter EH (2000). Heritable susceptibility for colitis in mice induced by IL-10 deficiency. Inflamm. Bowel Dis 6, 290–302. [DOI] [PubMed] [Google Scholar]
- Carvalho FA, Nalbantoglu I, Ortega-Fernandez S, Aitken JD, Su Y, Koren O, Walters WA, Knight R, Ley RE, Vijay-Kumar M, and Gewirtz AT (2012). Interleukin-1b (IL-1b) promotes susceptibility of Toll-like receptor 5 (TLR5) deficient mice to colitis. Gut 61, 373–384. [DOI] [PubMed] [Google Scholar]
- Cole JR, Wang Q, Fish JA, Chai B, McGarrell DM, Sun Y, Brown CT, Porras-Alfaro A, Kuske CR, and Tiedje JM (2014). Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Res 42, D633–D642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombel JF, Sandborn WJ, Rutgeerts P, Enns R, Hanauer SB, Panaccione R, Schreiber S, Byczkowski D, Li J, Kent JD, and Pollack PF (2007). Adalimumab for maintenance of clinical response and remission in patients with Crohn’s disease: the CHARM trial. Gastroenterology 132, 52–65. [DOI] [PubMed] [Google Scholar]
- Dixon P (2003). VEGAN, a package of R functions for community ecology. J. Veg. Sci 14, 927–930. [Google Scholar]
- Dubé PE, Yan F, Punit S, Girish N, McElroy SJ, Washington MK, and Polk DB (2012). Epidermal growth factor receptor inhibits colitis-associated cancer in mice. J. Clin. Invest 122, 2780–2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubé PE, Punit S, and Polk DB (2015). Redeeming an old foe: protective as well as pathophysiological roles for tumor necrosis factor in inflammatory bowel disease. Am. J. Physiol. Gastrointest. Liver Physiol 308, G161–G170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebach DR, Newberry R, and Stenson WF (2005). Differential role of tumor necrosis factor receptors in TNBS colitis. Inflamm. Bowel Dis 11, 533–540. [DOI] [PubMed] [Google Scholar]
- Eckhardt ER, Witta J, Zhong J, Arsenescu R, Arsenescu V, Wang Y, Ghoshal S, de Beer MC, de Beer FC, and de Villiers WJ (2010). Intestinal epithelial serum amyloid A modulates bacterial growth in vitro and pro-inflammatory responses in mouse experimental colitis. BMC Gastroenterol 10, 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer MA, Sundberg JP, Bristol IJ, Churchill GA, Li R, Elson CO, and Leiter EH (2001). A major quantitative trait locus on chromosome 3 controls colitis severity in IL-10-deficient mice. Proc. Natl. Acad. Sci. USA 98, 13820–13825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson LR, Han DY, Huebner C, Petermann I, Barclay ML, Gearry RB, McCulloch A, and Demmers PS (2009). Tumor necrosis factor receptor superfamily, member 1B haplotypes increase or decrease the risk of inflammatory bowel diseases in a New Zealand caucasian population. Gastroenterol. Res. Pract 2009, 591704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford AC, Sandborn WJ, Khan KJ, Hanauer SB, Talley NJ, and Moayyedi P (2011). Efficacy of biological therapies in inflammatory bowel disease: systematic review and meta-analysis. Am. J. Gastroenterol 106, 644–659, quiz 660. [DOI] [PubMed] [Google Scholar]
- Geirnaert A, Calatayud M, Grootaert C, Laukens D, Devriese S, Smagghe G, De Vos M, Boon N, and Van de Wiele T (2017). Butyrate-producing bacteria supplemented in vitro to Crohn’s disease patient microbiota increased butyrate production and enhanced intestinal epithelial barrier integrity. Sci. Rep 7, 11450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glocker E, and Grimbacher B (2012). Inflammatory bowel disease: is it a primary immunodeficiency? Cell. Mol. Life Sci 69, 41–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasberger H, Gao J, Nagao-Kitamoto H, Kitamoto S, Zhang M, Kamada N, Eaton KA, El-Zaatari M, Shreiner AB, Merchant JL, et al. (2015). Increased expression of DUOX2 is an epithelial response to mucosal dysbiosis required for immune homeostasis in mouse intestine. Gastroenterology 149, 1849–1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz R, Becker S, Sokolowski N, Schumann M, Bass D, and Malnick SD (2002). Murine monoclonal anti-tNF antibody administration has a beneficial effect on inflammatory bowel disease that develops in IL-10 knockout mice. Dig. Dis. Sci 47, 1723–1727. [DOI] [PubMed] [Google Scholar]
- Hale LP, and Greer PK (2012). A novel murine model of inflammatory bowel disease and inflammation-associated colon cancer with ulcerative colitis-like features. PLoS ONE 7, e41797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, et al. ; International IBD Genetics Consortium (IIBDGC) (2012). Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491, 119–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappelman MD, Moore KR, Allen JK, and Cook SF (2013). Recent trends in the prevalence of Crohn’s disease and ulcerative colitis in a commercially insured US population. Dig. Dis. Sci 58, 519–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzenik J, Larsen MD, Nielsen J, Kjeldsen J, and Nørgård BM (2019). Increased risk of developing Crohn’s disease or ulcerative colitis in 17 018 patients while under treatment with anti-TNFa agents, particularly etanercept, for autoimmune diseases other than inflammatory bowel disease. Aliment. Pharmacol. Ther 50, 289–294. [DOI] [PubMed] [Google Scholar]
- Kotlarz D, Beier R, Murugan D, Diestelhorst J, Jensen O, Boztug K, Pfeifer D, Kreipe H, Pfister ED, Baumann U, et al. (2012). Loss of interleukin-10 signaling and infantile inflammatory bowel disease: implications for diagnosis and therapy. Gastroenterology 143, 347–355. [DOI] [PubMed] [Google Scholar]
- Kühn R, Löhler J, Rennick D, Rajewsky K, and Müller W (1993). Interleukin-10-deficient mice develop chronic enterocolitis. Cell 75, 263–274. [DOI] [PubMed] [Google Scholar]
- Kullberg MC, Rothfuchs AG, Jankovic D, Caspar P, Wynn TA, Gorelick PL, Cheever AW, and Sher A (2001). Helicobacter hepaticus-induced colitis in interleukin-10-deficient mice: cytokine requirements for the induction and maintenance of intestinal inflammation. Infect. Immun 69, 4232–4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kullberg MC, Jankovic D, Feng CG, Hue S, Gorelick PL, McKenzie BS, Cua DJ, Powrie F, Cheever AW, Maloy KJ, and Sher A (2006). IL-23 plays a key role in Helicobacter hepaticus-induced T cell-dependent colitis. J. Exp. Med 203, 2485–2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappalainen M, Halme L, Turunen U, Saavalainen P, Einarsdottir E, Färkkilä M, Kontula K, and Paavola-Sakki P (2008). Association of IL23R, TNFRSF1A, and HLA-DRB1*0103 allele variants with inflammatory bowel disease phenotypes in the Finnish population. Inflamm. Bowel Dis 14, 1118–1124. [DOI] [PubMed] [Google Scholar]
- Le Hir M, Bluethmann H, Kosco-Vilbois MH, Müller M, di Padova F, Moore M, Ryffel B, and Eugster HP (1996). Differentiation of follicular dendritic cells and full antibody responses require tumor necrosis factor receptor-1 signaling. J. Exp. Med 183, 2367–2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Silverberg MS, Haritunians T, Dubinsky MC, Landers C, Stempak JM, Milgrom R, Guo X, Chen YD, Rotter JI, et al. (2016). TNFRSF1B Is Associated with ANCA in IBD. Inflamm. Bowel Dis 22, 1346–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CY, Xiao C, Fraser SE, Lester HA, and Koos DS (2012). Electrophysiological characterization of Grueneberg ganglion olfactory neurons: spontaneous firing, sodium conductance, and hyperpolarization-activated currents. J. Neurophysiol 108, 1318–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mähler M, Most C, Schmidtke S, Sundberg JP, Li R, Hedrich HJ, and Churchill GA (2002). Genetics of colitis susceptibility in IL-10-deficient mice: backcross versus F2 results contrasted by principal component analysis. Genomics 80, 274–282. [DOI] [PubMed] [Google Scholar]
- Matsumoto M, Mariathasan S, Nahm MH, Baranyay F, Peschon JJ, and Chaplin DD (1996). Role of lymphotoxin and the type I TNF receptor in the formation of germinal centers. Science 271, 1289–1291. [DOI] [PubMed] [Google Scholar]
- Maukonen J, Kolho KL, Paasela M, Honkanen J, Klemetti P, Vaarala O, and Saarela M (2015). Altered fecal microbiota in paediatric inflammatory bowel disease. J. Crohn’s Colitis 9, 1088–1095. [DOI] [PubMed] [Google Scholar]
- McGovern DPB, Kugathasan S, and Cho JH (2015). Genetics of inflammatory bowel diseases. Gastroenterology 149, 1163–1176.e1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurdie PJ, and Holmes S (2013). phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 8, e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi E, Hachiya Y, Kawada M, Nagatani K, Ogawa A, Sugimoto K, Mizoguchi A, and Podolsky DK (2008). TNF receptor type I-dependent activation of innate responses to reduce intestinal damage-associated mortality. Gastroenterology 134, 470–480. [DOI] [PubMed] [Google Scholar]
- Mombaerts P, Mizoguchi E, Grusby MJ, Glimcher LH, Bhan AK, and Tonegawa S (1993). Spontaneous development of inflammatory bowel disease in T cell receptor mutant mice. Cell 75, 274–282. [DOI] [PubMed] [Google Scholar]
- Moran CJ, Walters TD, Guo CH, Kugathasan S, Klein C, Turner D, Wolters VM, Bandsma RH, Mouzaki M, Zachos M, et al. (2013). IL-10R polymorphisms are associated with very-early-onset ulcerative colitis. Inflamm. Bowel Dis 19, 115–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murch SH, Lamkin VA, Savage MO, Walker-Smith JA, and MacDonald TT (1991). Serum concentrations of tumour necrosis factor alpha in childhood chronic inflammatory bowel disease. Gut 32, 913–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A, Glickman JN, Siebert R, Baron RM, Kasper DL, and Blumberg RS (2012). Microbial exposure during early life has persistent effects on natural killer T cell function. Science 336, 489–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasparakis M, Alexopoulou L, Episkopou V, and Kollias G (1996). Immune and inflammatory responses in TNF alpha-deficient mice: a critical requirement for TNF alpha in the formation of primary B cell follicles, follicular dendritic cell networks and germinal centers, and in the maturation of the humoral immune response. J. Exp. Med 184, 1397–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer K, Matsuyama T, Kündig TM, Wakeham A, Kishihara K, Shahinian A, Wiegmann K, Ohashi PS, Krönke M, and Mak TW (1993). Mice deficient for the 55 kd tumor necrosis factor receptor are resistant to endotoxic shock, yet succumb to L. monocytogenes infection. Cell 73, 457–467. [DOI] [PubMed] [Google Scholar]
- Pié S, Lallès JP, Blazy F, Laffitte J, Sève B, and Oswald IP (2004). Weaning is associated with an upregulation of expression of inflammatory cytokines in the intestine of piglets. J. Nutr 134, 641–647. [DOI] [PubMed] [Google Scholar]
- Pierik M, Vermeire S, Steen KV, Joossens S, Claessens G, Vlietinck R, and Rutgeerts P (2004). Tumour necrosis factor-alpha receptor 1 and 2 polymorphisms in inflammatory bowel disease and their association with response to infliximab. Aliment. Pharmacol. Ther 20, 303–310. [DOI] [PubMed] [Google Scholar]
- Pigneur B, Escher J, Elawad M, Lima R, Buderus S, Kierkus J, Guariso G, Canioni D, Lambot K, Talbotec C, et al. (2013). Phenotypic characterization of very early-onset IBD due to mutations in the IL10, IL10 receptor alpha or beta gene: a survey of the Genius Working Group. Inflamm. Bowel Dis 19, 2820–2828. [DOI] [PubMed] [Google Scholar]
- Punit S, Dube PE, Liu CY, Girish N, Washington MK, and Polk DB (2015). Tumor necrosis factor receptor 2 restricts the pathogenicity of CD8+ T cells in mice with colitis. Gastroenterology 149, 993–1005.e1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redhu NS, Bakthavatchalu V, Conaway EA, Shouval DS, Tsou A, Goettel JA, Biswas A, Wang C, Field M, Muller W, et al. (2017). Macrophage dysfunction initiates colitis during weaning of infant mice lacking the interleukin-10 receptor. eLife 6, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadlack B, Merz H, Schorle H, Schimpl A, Feller AC, and Horak I (1993). Ulcerative colitis-like disease in mice with a disrupted interleukin-2 gene. Cell 75, 253–261. [DOI] [PubMed] [Google Scholar]
- Salzer E, Kansu A, Sic H, Majek P, Ikinciogullari A, Dogu FE, Prengemann NK, Santos-Valente E, Pickl WF, Bilic I, et al. (2014). Early-onset inflammatory bowel disease and common variable immunodeficiency-like disease caused by IL-21 deficiency. J. Allergy Clin. Immunol. 133, 1651–1659.e1612. [DOI] [PubMed] [Google Scholar]
- Sashio H, Tamura K, Ito R, Yamamoto Y, Bamba H, Kosaka T, Fukui S, Sawada K, Fukuda Y, Tamura K, et al. (2002). Polymorphisms of the TNF gene and the TNF receptor superfamily member 1B gene are associated with susceptibility to ulcerative colitis and Crohn’s disease, respectively. Immunogenetics 53, 1020–1027. [DOI] [PubMed] [Google Scholar]
- Scheer S, Medina TS, Murison A, Taves MD, Antignano F, Chenery A, Soma KK, Perona-Wright G, Lupien M, Arrowsmith CH, et al. (2017). Early-life antibiotic treatment enhances the pathogenicity of CD4+ T cells during intestinal inflammation. J. Leukoc. Biol 101, 893–900. [DOI] [PubMed] [Google Scholar]
- Scheinin T, Butler DM, Salway F, Scallon B, and Feldmann M (2003). Validation of the interleukin-10 knockout mouse model of colitis: antitumour necrosis factor-antibodies suppress the progression of colitis. Clin. Exp. Immunol 133, 38–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schildkraut V, Alex G, Cameron DJ, Hardikar W, Lipschitz B, Oliver MR, Simpson DM, and Catto-Smith AG (2013). Sixty-year study of incidence of childhood ulcerative colitis finds eleven-fold increase beginning in 1990s. Inflamm. Bowel Dis 19, 1–6. [DOI] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol 75, 7537–7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber S, Khaliq-Kareemi M, Lawrance IC, Thomsen OO, Hanauer SB, McColm J, Bloomfield R, and Sandborn WJ; PRECISE 2 Study Investigators (2007). Maintenance therapy with certolizumab pegol for Crohn’s disease. N. Engl. J. Med 357, 239–250. [DOI] [PubMed] [Google Scholar]
- Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, and Huttenhower C (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12, R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh V, Kumar M, San Yeoh B, Xiao X, Saha P, Kennett MJ, and Vijay-Kumar M (2016). Inhibition of interleukin-10 signaling induces microbiotadependent chronic colitis in apolipoprotein E deficient mice. Inflamm. Bowel Dis 22, 841–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surana NK, and Kasper DL (2017). Moving beyond microbiome-wide associations to causal microbe identification. Nature 552, 244–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegtmeyer D, Seidl M, Gerner P, Baumann U, and Klemann C (2017). Inflammatory bowel disease caused by primary immunodeficiencies−clinical presentations, review of literature, and proposal of a rational diagnostic algorithm. Pediatr. Allergy Immunol 28, 412–429. [DOI] [PubMed] [Google Scholar]
- Toussirot É, Houvenagel É, Goëb V, Fouache D, Martin A, Le Dantec P, Dernis E, Wendling D, Ansemant T, Berthelot JM, et al. ; Le CRI (2012). Development of inflammatory bowel disease during anti-TNF-α therapy for inflammatory rheumatic disease: a nationwide series. Joint Bone Spine 79, 457–463. [DOI] [PubMed] [Google Scholar]
- Üsküdar Cansu D, Üsküdar Teke H, Temel T, Ertürk A, Kahraman O, and Korkmaz C (2019). Do anti-TNF agents increase the risk of inflammatory bowel disease evolution in patients with ankylosing spondylitis? real life data. J. Natl. Med. Assoc 111, 262–269. [DOI] [PubMed] [Google Scholar]
- Vital M, Howe AC, and Tiedje JM (2014). Revealing the bacterial butyrate synthesis pathways by analyzing (meta)genomic data. MBio 5, e00889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walthall K, Cappon GD, Hurtt ME, and Zoetis T (2005). Postnatal development of the gastrointestinal system: a species comparison. Birth Defects Res. B Dev. Reprod. Toxicol 74, 132–156. [DOI] [PubMed] [Google Scholar]
- Wang K, Han G, Dou Y, Wang Y, Liu G, Wang R, Xiao H, Li X, Hou C, Shen B, et al. (2012). Opposite role of tumor necrosis factor receptors in dextran sulfate sodium-induced colitis in mice. PLoS ONE 7, e52924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Han G, Chen Y, Wang K, Liu G, Wang R, Xiao H, Li X, Hou C, Shen B, et al. (2013). Protective role of tumor necrosis factor (TNF) receptors in chronic intestinal inflammation: TNFR1 ablation boosts systemic inflammatory response. Lab. Invest 93, 1024–1035. [DOI] [PubMed] [Google Scholar]
- Waschke KA, Villani AC, Vermeire S, Dufresne L, Chen TC, Bitton A, Cohen A, Thomson AB, and Wild GE (2005). Tumor necrosis factor receptor gene polymorphisms in Crohn’s disease: association with clinical phenotypes. Am. J. Gastroenterol 100, 1126–1133. [DOI] [PubMed] [Google Scholar]
- Yu YR, O’Koren EG, Hotten DF, Kan MJ, Kopin D, Nelson ER, Que L, and Gunn MD (2016). A protocol for the comprehensive flow cytometric analysis of immune cells in normal and inflamed murine non-lymphoid tissues. PLoS ONE 11, e0150606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Dubé PE, Washington MK, Yan F, and Polk DB (2012). ErbB2 and ErbB3 regulate recovery from dextran sulfate sodium-induced colitis by promoting mouse colon epithelial cell survival. Lab. Invest 92, 437–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq datasets generated during this study are available at NCBI Gene Expression Omnibus (accession numbers): GSE107933, GSE155654, GSE155626).


