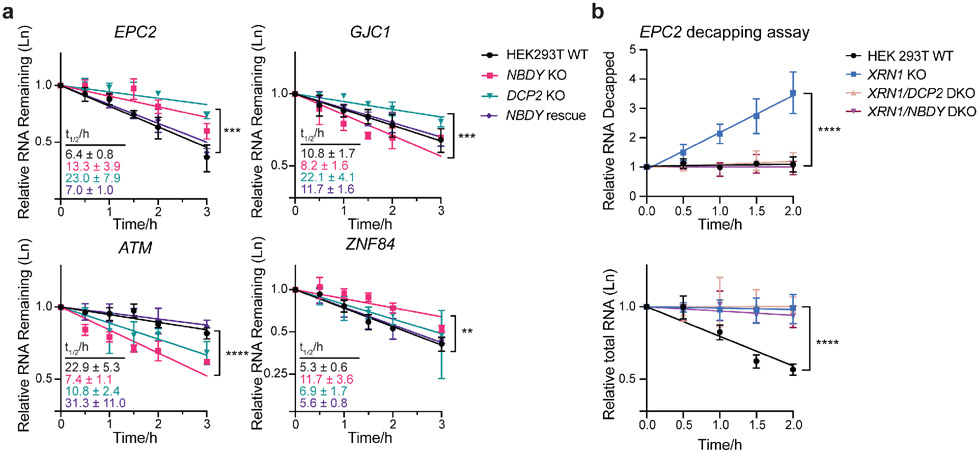
Figure 6∣. NBDY regulates stability of DCP2 target transcripts at the decapping step.
(a) RNA stability measurement of selected genes belonging to the following classes: stabilized in both DCP2 KO and NBDY KO (EPC2), stabilized by DCP2 KO but destabilized in NBDY KO (GJC1), and destabilized in both DCP2 KO and NBDY KO (ATM), stabilized in NBDY KO and unaffected in DCP2 KO (ZNF84). Number of biological replicates: n=3. Error bars represent mean ± s.d. Significance was analyzed by ANOVA linear regression. **P < 0.01; ***P < 0.001; ****P < 0.0001, Dunnett’s test. (b) Splinted ligation RT-PCR (qSL-RT-PCR) in WT, XRN1 KO, XRN1/DCP2 or XRN1/NBDY double knock out (DKO) HEK 293T cell lines was performed to assay decapping of a transcript stabilized by both DCP2 and NBDY KO, EPC2 (top). Total RNA was quantified by qRT-PCR at each timepoint (bottom). Significance was analyzed by ANOVA linear regression. Error bars represent mean ± s.d. n=4 biological replicates. ****P < 0.0001, Dunnett’s test.