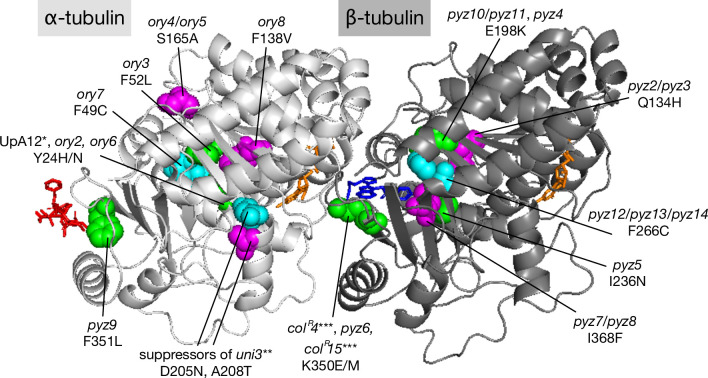
Fig 5. Predicted three-dimensional structure of Chlamydomonas reinhardtii α/β-tubulin heterodimer showing the mutations reported in the present and previous studies.
Light gray, α-tubulin; dark gray, β-tubulin. Altered amino acids are shown as sphere representations. The binding sites of tubulysin M (red, an oryzalin-like compound) and 2RR (blue, 3-[(4-{1-[2-(4-aminophenyl)-2-oxoethyl]-1H-benzimidazol-2-yl}-1,2,5-oxadiazol-3-yl)amino]propanenitrile, a propyzamide-like compound) were determined by applying the alignment command in MacPyMol software to the tubulin structures 4ZOL and 4O2A reported in the presence of these compounds [25, 26]. The orange stick representations show GTP (in α-tubulin) and GDP (in β-tubulin). Mutations identical to previously reported mutations are marked with asterisks: *, [3]; **, [5]; and ***, [6].