Abstract
Error-free cell division depends on the accurate assembly of the spindle midzone from dynamic spindle microtubules to ensure chromatid segregation during metaphase–anaphase transition. However, the mechanism underlying the key transition from the mitotic spindle to central spindle before anaphase onset remains elusive. Given the prevalence of chromosome instability phenotype in gastric tumorigenesis, we developed a strategy to model context-dependent cell division using a combination of light sheet microscope and 3D gastric organoids. Light sheet microscopic image analyses of 3D organoids showed that CENP-E inhibited cells undergoing aberrant metaphase–anaphase transition and exhibiting chromosome segregation errors during mitosis. High-resolution real-time imaging analyses of 2D cell culture revealed that CENP-E inhibited cells undergoing central spindle splitting and chromosome instability phenotype. Using biotinylated syntelin as an affinity matrix, we found that CENP-E forms a complex with PRC1 in mitotic cells. Chemical inhibition of CENP-E in metaphase by syntelin prevented accurate central spindle assembly by perturbing temporal assembly of PRC1 to the midzone. Thus, CENP-E-mediated PRC1 assembly to the central spindle constitutes a temporal switch to organize dynamic kinetochore microtubules into stable midzone arrays. These findings reveal a previously uncharacterized role of CENP-E in temporal control of central spindle assembly. Since CENP-E is absent from yeast, we reasoned that metazoans evolved an elaborate central spindle organization machinery to ensure accurate sister chromatid segregation during anaphase and cytokinesis.
Keywords: organoids, cell division, central spindle, CENP-E, syntelin, PRC1
Introduction
Organism physiology requires accurate cell division and renewal. Chromosome segregation errors can lead to chromosome damage and aneuploidy that are prevalent in cancer. During cell division cycle, accurate chromosome segregation in mitosis depends on the spindle assembly checkpoint that prevents premature exit from mitosis (Cleveland et al., 2003; McKinley et al., 2018). In addition, chromosome segregation error can also trigger cell death progress to avoid the inflammatory chain response and harboring chromosome instability in tissue (Bakhoum et al., 2018). Congression of chromosomes to the metaphase plate is driven by a processive, plus end-directed kinetochore motor CENP-E (Yao et al., 2000; Kapoor et al., 2006; Kim et al., 2010). During prometaphase–metaphase transition, the motility of CENP-E converts from a lateral mode into an end-on mode, and it maintains its association with both the assembling and disassembling microtubule plus ends during chromosome oscillation (Gudimchuk et al., 2013). CENP-E exhibits a dynamic distribution from kinetochore to the midzone of central spindle during metaphase–anaphase transition (Yao et al., 1997; Dou et al., 2015; Ding et al., 2019; Huang et al., 2019). However, the mechanisms underlying the switch of CENP-E from mitotic spindle to central spindle remain unknown.
Microtubule plus-end tracking proteins (+TIPs), a large group of evolutionarily unrelated proteins, bind selectively to growing microtubule plus ends (Akhmanova and Steinmetz, 2008; Liu et al., 2009; Ward et al., 2013; Xia et al., 2014; Adams et al., 2016; Huang et al., 2019). +TIPs have been shown to be involved in regulating microtubule dynamics and chromosome segregation. PRC1, a different group of proteins, selectively binds to overlapping microtubules, thus forming the central spindle (or the midzone) during cell division (Fu et al., 2007; Liu et al., 2009; Subramanian et al., 2013). However, it remains elusive as how PRC1 targets to the central spindle during metaphase–anaphase transition and whether PRC1 cooperates with kinetochore molecules such as CENP-E to switch spindle microtubule to central spindle.
Recent studies show that epithelial cells exhibit high chromosome segregation fidelity in tissues but that this is lost when tissue 3D structure is disrupted (Knouse et al., 2018; McKinley et al., 2018). The stomach shares a number of features with the intestine, including a common endodermal origin and a constantly renewing epithelium (Forte and Yao, 1996; Yao and Smolka, 2019). The renewal of gastrointestinal epithelial cells is fueled from stem cells populations located within the epithelium. In the stomach, the epithelium is organized into multiple gastric units that are comprised of flask-shaped tubular glands, several of which feed into a single pit that opens out onto the surface epithelium (Yao and Forte, 2003; Yao and Smolka, 2019). These multi-potent stem cells, which are responsible for the renewal of the gastric epithelium, enabled the establishment of long-lived organoids resembling mature pyloric epithelium in the stomach (Barker et al., 2010). Each gastric organoids are considered to be functionally monoclonal, with all cellular progeny being derived from a single stem cell (McDonald et al., 2008; McCracken et al., 2014; Bartfeld et al., 2015; Eicher et al., 2018). This dominant stem cell is thought to maintain a small, steady-state population of clonal, multi-potent stem cells in the gastric unit through infrequent symmetric division. Daughter cells generated by much more frequent asymmetric division of these multi-potent stem cells subsequently exit the stem cell niche and differentiate to generate the parietal and chief cell lineage as they migrate bi-directionally toward the gland (Bjerknes and Cheng, 2002). Thus, we consider gastric organoids as a unique model system to illuminating the cellular dynamics underlying stem cell division and plasticity control with respect to specific contexts in physiology and in diseases.
Here, we report modeling physiology of cell division control in epithelial cells using 3D gastric organoids and chemical inhibitors of mitosis. Remarkably, two structurally distinct CENP-E chemical inhibitors syntelin and GSK923295 perturb cell metaphase–anaphase transition by antagonizing central spindle assembly. Syntelin does not perturb inter-polar microtubule assembly but abrogates the anti-parallel microtubule bundle formation. Using syntelin as an affinity matrix coupled to mass spectrometric identification of proteins bound to syntelin, we have identified a novel protein complex containing CENP-E–PRC1. Importantly, the interaction between CENP-E and PRC1 establishes stable midzone arrays for accurate central spindle assembly. Our results highlight the strength of modeling mitosis in organoids as it faithfully recapitulates fundamental cellular processes.
Results
Insights from modeling of mitosis in 3D gastric organoids
Gastric cancer is a leading cause of cancer deaths; the Cancer Genome Atlas (TCGA) project proposed a molecular classification dividing gastric cancer into four subtypes with common characteristics on chromosomal instability, which provides a roadmap for design of targeted therapies of gastric cancer. An emerging therapeutic strategy for cancer is to induce selective lethality in a tumor by exploiting interactions between its driving mutations and specific drug targets. To this end, we established a screening platform, including FDA-approved drugs for synthetic lethality in the clinic oncology (data not shown) and mitotic inhibitors for delineating mechanisms of action underlying chromosomal instability. Based on this platform, we imaged cellular dynamics of human gastric organoids derived from gastric carcinoma, which carry distinct genetic mutation, in response to those chemical compounds (Figure 1A). Typically, the gastric organoids were generated as described (Barker et al., 2010) and have been successfully replated on a weekly basis for at least 3 months, without losing the properties such as histamine-elicited acid secretion. Gastric organoids typically appeared as hollow structures with a sealed lumen surrounded by a single layer of cylindrical epithelial cells under light sheet microscope, visualized by staining with filamentous actin marker phalloidin and 4’,6-diamidino-2-phenylindole (DAPI; Supplementary Figure S1B).
Figure 1.
Modeling mitosis using a light sheet microscopy and chemical probes. (A) Cartoon presentation of generation of patient-derived gastric organoids. The organoid model provides a unique platform to delineate cellular dynamics during cell division in native tissue and imply synthetic lethal strategy to identify individualized medication. (B) 3D projections of gastric organoids montages from four different focal planes (Z = 92, 126, 142, and 246, respectively) that were labeled with tubulin and DAPI. The magnified images provide apparent configuration of chromosome relative to mitotic spindle. The lower imaging toxicity combined with unlimited sample depth has established light sheet microscope as a unique platform for studying molecular and cellular dynamics in live tissue. Aberrant chromosome movements such as misalignments are indicated by arrows. Scale bar, 100 μm in organoids imaging; 10 μm in magnified montage of individual mitotic cells. (C) Statistical analyses of mitotic index from different chemical probes such as CENP-E inhibitors. Note that CENP-E inhibition by syntelin and GSK923295 exhibits chronic mitotic arrest. Data represent mean ± SEM from three independent experiments. Statistical significance was determined by two-sided t-test (**P < 0.01; n = 21 for each group). (D) Magnified montage of an anaphase cells treated with syntelin. The magnified anaphase cell exhibits typical lagging chromosome (arrow). (E) Statistical analyses of aberrant anaphase cells from organoids treated with mitotic modulators such as CENP-E inhibitors. Note that CENP-E inhibition by syntelin and GSK923295 significantly increased the number of cells exhibiting aberrant anaphase chromosomes (**P < 0.01; n = 23 for each group). Data represent mean ± SEM from three independent experiments.
To probe the cellular dynamics of mitosis in 3D environment, we treated gastric organoids with various chemical modulators of mitotic regulators such as BI2536 (PLK1 inhibitor; Steegmaier et al., 2007), GSK923295 (CENP-E kinesin inhibitor; Wood et al., 2010), Syntelin (CENP-E kinesin inhibitor; Ding et al., 2010), and monastrol (EG5 kinesin inhibitor; Mayer et al., 1999) for 60 min to visualize the mitotic phenotype associated with inhibition of mitotic kinase or kinesin (Supplementary Figure S1A). To determine whether the stem cell division arrest that arises from inhibition of CENP-E reflects an underlying defect in spindle assembly or maintenance, we visualized spindles with antibodies against tubulin and identified chromosomes with DAPI. As expected, control cells in dimethyl sulfoxide (DMSO)-treated organoids passed through the mitosis with all phases of the mitotic cells (including early and late prometaphase, metaphase, and early and late anaphase, respectively; Supplementary Figure S1B). In contrast, at comparable time points, most CENP-E inhibited cells had established bipolar spindles but each contained misaligned and lagging chromosomes that were readily apparent in the enlarged montages (Figure 1B). This syntelin-elicited mitotic arrest phenotype was also apparent in GSK923295-treated organoids (Supplementary Figure S1C). Since GSK923295 exhibits completely different structure of syntelin but shares common inhibition on CENP-E, the mitotic arrest phenotype in syntelin-treated cells indicates the importance of functional CENP-E in cell division in gastric organoids. Statistical analyses indicate that modeling of mitosis using chemical inhibitors of CENP-E, PLK1 etc. is efficient as CENP-E and PLK1 inhibitors specifically arrest gastric stem cells in mitosis in gastric organoids (Figure 1C). Additional phenotype-based screen with Eg5 inhibitor monastrol and PLK1 inhibitor BI2536 revealed expected mitotic phenotype of monopolar spindle without perturbation of cell polarity of interphase cells (Supplementary Figure S1D and E), indicating cell–cell interactions, cell division circadian control, and 3D glandular architecture are maintained in gastric organoids. Thus, we conclude that 3D gastric organoids is a physiological model system to study cell division control in response to the dynamic changes of intracellular and extracellular cues.
To systemically examine the mitotic phenotypes associated with chemical inhibition, we carried out examination of serial sections of treated organoids. Careful examination of serial section of gastric organoids revealed that premature anaphase hallmarked by the chromosome bridges between partially separated sister chromatids (dashed box in Figure 1B, bottom panel; Z = 246). At a higher magnification, one premature anaphase cell with the lagging chromosomes at the midzone of the separating sister chromatids was imaged (Figure 1D, arrow). Careful examination of mitotic cells from three different preparation indicated that syntelin and GSK923295 arrest cells in premature anaphase (Figure 1E). As a control, Mps1 kinase inhibitor (reversine) treatment did not significantly increase the percentage of aberrant anaphase cells. Thus, we conclude that CENP-E is essential for accurate mitosis and healthy cell renewal in organoids.
CENP-E motor activity is essential for accurate anaphase onset
The functional relevance of CENP-E in metaphase–anaphase transition during mitosis has been suggested from an early antibody injection experiment in which microinjection of the mAb177 into metaphase cells blocks or significantly delays progression into anaphase (Yen et al., 1991). However, the precise role and underlying mechanism have never been directly examined due to the lack of analytical tools for temporal dissection. In addition, the classical tools such as siRNA-mediated suppression do not distinguish the enzymatic function of kinesin from its structural role. To characterize that the premature anaphase phenotype seen in syntelin-treated cells from gastric organoids is not a tissue-specific feature, we took the advantage of high-efficient cell cycle synchronization in 2D culture of HeLa cells (Liu et al., 2009; Ding et al., 2010). As schematically illustrated in Figure 2A, HeLa cells were synchronized in metaphase using monastrol plus MG132 treatment followed by released into syntelin treatment to see whether CENP-E is required for anaphase onset. To examine the precise function of CENP-E, HeLa cells expressing GFP-H2B and mCherry-tubulin were treated with monastrol and MG132 to synchronize HeLa cells in metaphase (Mo et al., 2016). After three washes with Dulbecco’s modified Eagle’s medium (DMEM) media containing DMSO and syntelin (1 μM) but without MG132, HeLa cells were then subjected to real-time imaging as previously reported (Ding et al., 2010). As shown in Figure 2B, we began real-time imaging of metaphase-synchronized HeLa cells expressing mCherry-tubulin and GFP-H2B to visualize chromosome alignment and subsequent metaphase–anaphase transition. In general, it takes an average of 50 ± 4 min (n = 8 cells) for HeLa cells to transit from metaphase to the anaphase onset of sister chromatid separation. Although the average time (54 ± 4 min; n = 8 cells) for syntelin-treated cells to transit from metaphase to anaphase onset is similar to that of DMSO-treated cells, careful examination revealed that the sister chromatids were not equally segregated into anaphase as some of the chromatids were further split (Figure 2C, lower panel, white arrowheads). Careful examination of mitotic cells from three different preparation indicated that addition of syntelin and GSK923295 into metaphase cells perturbed accurate anaphase onset (Figure 2D). As a positive control, PLK1 kinase inhibitor (BI2536) treatment also exhibited less but significant perturbation of anaphase onset. Thus, we conclude that CENP-E is essential for metaphase–anaphase transition.
Figure 2.
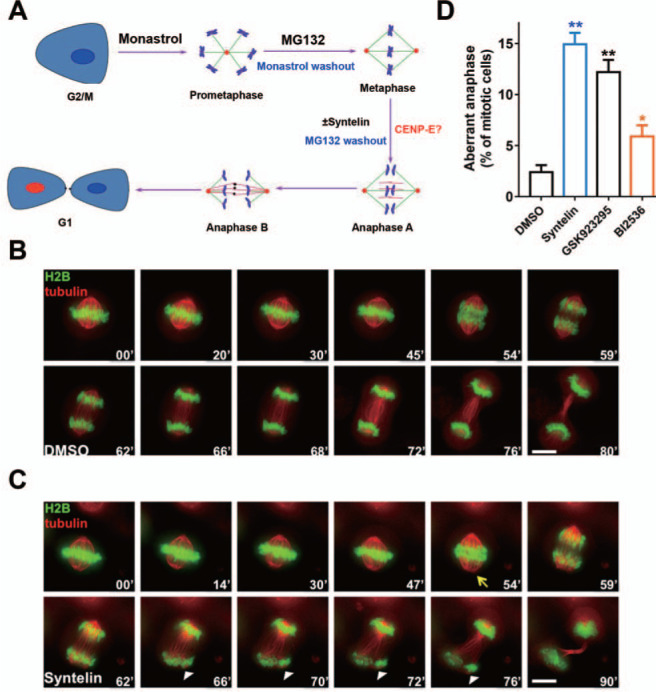
CENP-E kinesin activity is essential for accurate anaphase onset. (A) Schematic presentation of delineating temporal function of CENP-E in mitosis. HeLa cells were synchronized at metaphase using chemical inhibitors and exposed to syntelin treatment. (B) Representative montage from live HeLa cells expressing GFP-H2B and mCherry-tubulin. HeLa cells were transfected to express GFP-H2B and mCherry-tubulin for 24 h prior to MG132 synchronization and real-time imaging experimentation as illustrated in A. Note that the cell was treated with DMSO. Scale bar, 10 μm. (C) Representative montage from live HeLa cells expressing GFP-H2B and mCherry-tubulin with syntelin treatment. HeLa cells were transfected to express GFP-H2B and mCherry-tubulin for 24 h prior to MG132 synchronization and parallel experimentation with syntelin as in B. Note that the cell treated with 1 μM syntelin at the metaphase exhibited aberrant metaphase–anaphase transition with chromatid split phenotype. Note that arrowheads indicate the chromatid split phenotype. Scale bar, 10 μm. (D) Quantification of aberrant anaphase phenotypes of live HeLa cells treated with DMSO, syntelin (1 μM), GSK923295 (100 nM), or BI2536 (100 nM). Data represent mean ± SEM from three independent experiments. Statistical significance was tested by two-sided t-test (*P < 0.05; **P < 0.01; n = 25 for each group).
CENP-E is required to organize PRC1 to the spindle midzone during mitosis
The syntelin-elicited perturbation of central spindle morphology is reminiscent of what was seen in PRC1 impaired cells (Fu et al., 2007; Liu et al., 2009). CENP-E is a mitotic kinesin that accumulates in G2/M and translocates to kinetochore upon the nuclear envelope breakdown (Yen et al., 1991). During mitosis, CENP-E relocates from kinetochore to the central spindle during metaphase–anaphase transition (Yen et al., 1991). Immuno-electron microscopic analyses revealed that CENP-E translocates to the midzone during metaphase–anaphase transition and cross-links the interzonal microtubules beginning in anaphase B (Yao et al., 1997). We sought to examine whether PRC1 distributes with CENP-E to the midzone of mitotic cells. As shown in Supplementary Figure S2A, PRC1 signal is readily evident at the central spindle where it is superimposed to that of CENP-E (merge, ‘yellow fibers’) in anaphase cell. The co-distribution profile of CENP-E and PRC1 to the interzonal microtubules remains through telophase (merge, lower panel). As expected, exogenously expressed mCherry-PRC1 indeed colocalized with CENP-E to the central spindle and their co-distribution remains through telophase (Supplementary Figure S2B, ‘magenta fibers’). Thus, we conclude that PRC1 colocalizes with CENP-E to central spindle from anaphase to telophase during mitosis.
To examine whether CENP-E inhibition in metaphase alters the assembly of PRC1, we began real-time imaging of metaphase-synchronized HeLa cells expressing GFP-PRC1 and mCherry-H2B. Syntelin was added into prometaphase cells during the monastrol washout, and time-lapse imaging was started as the cells in prometaphase. In general, it takes an average of 54 ± 4 min (n = 8 cells) for control HeLa cells to transit from prometaphase to anaphase onset and PRC1-labeled fibers are well organized (Figure 3A). The GFP-PRC1-marked central spindle was disorganized as the syntelin-treated cells struggled to enter anaphase (Figure 3B; between 54 and 69 min), suggesting that CENP-E activity is required to organize PRC1 to the spindle midzone.
Figure 3.

CENP-E kinesin is required to organize PRC1 to the spindle midzone. (A) Representative montage from live HeLa cells expressing dual-color reporters (GFP-PRC1 and mCherry-H2B). HeLa cells were transfected to express GFP-PRC1 and mCherry-H2B for 24 h prior to synchronization at the metaphase for the real-time experimentation. Note that the cell treated with DMSO progressed into anaphase with accurate chromatid segregation. (B) Representative montage from live HeLa cells expressing dual-color reporters (GFP-PRC1 and mCherry-H2B) with syntelin treatment. HeLa cells were transfected to express GFP-PRC1 and mCherry-H2B for 24 h prior to synchronization at the metaphase for syntelin treatment and the real-time experimentation. Note that the cell treated with 1 μM syntelin at the metaphase exhibited perturbation in establishment of central spindle midzone and errors in sister chromatid separation and segregation. (C) Representative montage from live HeLa cells expressing GFP-PRC1. Note that PRC1-labeled central spindle was organized and dynamically concentrated into the midbody in DMSO-treated cells. (D) Representative montage from live HeLa cells expressing GFP-PRC1. Note that the cell was treated with 1 μM syntelin at the metaphase and syntelin treatment inhibited the formation of PRC1-organized anti-parallel central spindle. (E) Quantification of mitotic phenotypes of live HeLa cells treated with DMSO, syntelin (1 μM), GSK923295 (100 nM), or reversine (300 nM). Data represent mean ± SEM from three independent experiments. Statistical significance was tested by two-sided t-test (**P < 0.01; n = 50).
In a separate experiment using single color GFP-PRC1 for real-time analysis, we confirmed that syntelin treatment inhibited the formation of PRC1-organized anti-parallel central spindle as GFP-PRC1 hardly formed organized fibers in syntelin-treated cells (Figure 3D). Syntelin was added into metaphase cells during the MG132 washout, and time-lapse imaging was started as the cells underwent metaphase–anaphase transition. In the DMSO-treated cells, PRC1-labeled central spindle was organized and dynamically concentrated into the midbody (Figure 3C). Statistical analyses indicate midzonal microtubules and PRC1 organization are perturbed by CENP-E inhibitors syntelin and GSK923295, but not Mps1 inhibitor reversine (Figure 3E). Thus, we conclude that CENP-E motor activity is essential for accurate organization of PRC1-rich central spindle midzone during mitosis.
CENP-E is essential for accurate central spindle establishment
Our early immuno-electron microscopic analyses demonstrated that CENP-E was only found microtubule-associated in the electron dense region with overlapping microtubule plus ends (Yao et al., 1997), suggesting that CENP-E is located to stable anti-parallel microtubule midzone at metaphase–anaphase transition onset. To explore the potential role of CENP-E interaction in metaphase–anaphase transition, we synchronized HeLa cells at metaphase by MG132 followed by washout to allow cells to progress into anaphase in the presence or absence of syntelin as illustrated in Figure 2A. Syntelin-treated and control cells were then fixed and stained with CENP-E, tubulin, and DAPI, respectively. Careful examination of the microtubule network revealed that inhibition of CENP-E perturbed the establishment of a stable central spindle as syntelin-treated cells exhibited no apparent anti-parallel microtubules at anaphase onset (Figure 4A, lower panel, arrow). The central spindle in DMSO-treated cells is organized and apparent, suggesting that anti-parallel microtubules may not be established and/or stabilized in the absence of CENP-E motor activity.
Figure 4.
CENP-E kinesin is essential for accurate establishment of central spindle. (A) HeLa cells treated with DMSO and syntelin (1 μM) were subjected to immunofluorescence assay for CENP-E (red), tubulin (green), and DAPI. Note that syntelin-treated cells exhibit aberrant central spindle revealed by tubulin staining (arrow). Scale bar, 10 μm. (B) HeLa cells treated with DMSO and syntelin (1 μM) were subjected to immunofluorescence assay for MKLP1 (red), tubulin (green), and DAPI. Note that syntelin-treated cells exhibit virtually no MKLP1 (arrow). Scale bar, 10 μm. (C) Quantification of central spindle location of CENP-E, MKLP1, and PRC1 in HeLa cells treated with DMSO, syntelin (1 μM), GSK923296 (100 nM), and reversine (300 nM), respectively. Data represent mean ± SEM from three independent experiments. Statistical significance was tested by two-sided t-test (**P < 0.01; n = 50). (D) Inhibition of CENP-E perturbed central spindle organization. HeLa cells were processed as described in Materials and methods prior to electron microscopic analyses of anaphase central spindle of HeLa cells treated with DMSO and syntelin (1 μM). (a) Lower magnification of view of a late anaphase HeLa cell bearing elongated spindle poles, labeled with asterisks. Interzonal microtubules are readily seen (boxed). Scale bar, 2 μm. (b) Magnified view of the boxed region in a, showing that anti-parallel microtubules are readily apparent (red arrowhead). Arrows indicate CENP-E gold particle deposition. Scale bar, 200 nm. (c) Lower magnification of view of a late anaphase HeLa cell treated with syntelin, labeled with asterisks. Interzonal microtubules are perturbed without apparent central spindle (boxed). Scale bar, 2 μm. (d) Magnified view of the boxed region in c, showing that anti-parallel microtubules are collapsed (red arrow). Scale bar, 200 nm. Note that syntelin treatment prevents the establishment of anti-parallel microtubules that fail to power the elongation of central spindles. (E) Schematic presentation of working model to account for CENP-E function in central spindle anti-parallel microtubule establishment and organization. We proposed that CENP-E organizes anti-parallel microtubules and powers microtubule sliding in overlapped midzone.
To score the impact of CENP-E inhibition on central spindle assembly, we next examined the location of MKLP1, a critical kinesin for central spindle elongation as anaphase progression. To this end, syntelin-treated and control cells were then fixed and stained with MKLP1, tubulin, and DAPI, respectively. Interestingly, inhibition of CENP-E by chemical inhibitor syntelin prevented central spindle location of MKLP1 as no apparent MKLP1 molecules are located to the central spindle in syntelin-treated cells (Figure 4B, bottom panel, arrow). On the other hand, the central spindle localization of MKLP1 in DMSO-treated cells is organized and apparent, suggesting that CENP-E motor activity is important for localization of central spindle organizers such as MKLP1. Statistical analyses indicate that CENP-E is essential for organization of midzonal microtubule and localization of MKLP1 (Figure 4C).
Error-free cell division depends on the accurate and dynamic reorganization of the spindle midzone from spindle microtubules during metaphase–anaphase onset. To determine whether inhibition of CENP-E prevents anti-parallel microtubule establishment, we carried out immuno-electron microscopic analyses of HeLa cells treated with syntelin and DMSO. As shown in Figure 4D, anti-parallel microtubules are readily apparent in DMSO-treated cells in a magnified montage (upper right panel). The 10-nm gold particles are surrounding the anti-parallel microtubule bundles, which demonstrated that CENP-E motor activity is critical for stable anti-parallel microtubule midzone establishment. Significantly, no apparent anti-parallel microtubules are found in syntelin-treated anaphase cell (Figure 4D, lower right panel). Thus, we conclude that CENP-E is essential for accurate organization of anti-parallel central spindles (Figure 4E).
CENP-E forms a functional complex with PRC1 in mitosis
To delineate the mechanism underlying syntelin-elicited aberrant central spindle organization, we sought to examine whether there is a temporally regulated complex of CENP-E with PRC1 in metaphase–anaphase transition. We took the advantage of CENP-E inhibitor syntelin as a functional linker and used biotinylated syntelin as an affinity matrix to isolate CENP-E interacting proteins as illustrated in Figure 5A. Our trial experiment showed that biotinylated syntelin can generate characteristic chromosome with syntelic attachment in a similar manner to that of unconjugated syntelin (Supplementary Figure S3), suggesting that biotinylation did not alter syntelin binding to CENP-E. After demonstrating the labeling efficiency of biotinylated syntelin, we used avidin-sepharose beads as affinity matrix to absorb CENP-E and its binding partners bound to biotinylated syntelin from mitotic cell lysates. After extensive washing, unconjugated syntelin was used to elute proteins bound to biotinylated syntelin. The resulting pull-down proteins from syntelin competition were subjected to sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) analyses followed by Coomassie Brilliant Blue staining and mass spectrometric identification (Figure 5B). Using western blotting analyses, we confirmed that CENP-E, BubR1, and PRC1 proteins were recovered from syntelin affinity pull-down assay (Figure 5C, lane 2). These analyses indicate that CENP-E forms a complex with PRC1 in mitotic cells.
Figure 5.
CENP-E forms a functional complex with PRC1 in mitosis. (A) Schematic drawing of syntelin-affinity matrix-based discovery of CENP-E binders. In brief, clarified mitotic HeLa cell lysates were subjected to binding to biotinylated syntelin followed by absorption with avidin-sepharose beads. The bound materials after extensive wash will be resolved by SDS-PAGE followed by mass spectrometric identification. (B) Representative SDS-PAGE analyses from syntelin-affinity purification. The clarified mitotic HeLa cell lysates were used as inputs for incubation of biotinylated syntelin followed by isolation with avidin-based affinity matrix. The proteins bound to avidin matrix were eluted with syntelin and resolved by SDS-PAGE followed by Coomassie Brilliant Blue staining. Mass spectrometric analyses indicate that CENP-E and PRC1 were recovered from avidin matrix. (C) Representative western blotting analyses for proteins isolated by syntelin-affinity purification. Note that BubR1, CENP-E, and PRC1 were isolated by syntelin–biotin–avidin matrix. (D) Identification of PRC1 as a cognate binding protein of CENP-E. The clarified mitotic HeLa cell lysates were used as inputs for incubation of anti-CENP-E antibodies followed by Protein A/G affinity chromatography. Immunoprecipitation experiment showed that endogenous PRC1 binds to CENP-E. (E) Working model accounts for novel CENP-E function in facilitating an initial central spindle establishment via a temporally controlled CENP-E–PRC1 interaction. CENP-E is situated on the outermost surface of the kinetochore during initial lateral microtubule attachment. After biorientation, CENP-E links kinetochores to spindle microtubules to achieve metaphase alignment. At metaphase, CENP-E releases and redistributes to midzone with PRC1 where they organize interzonal microtubules and power microtubule sliding that leads to pole-to-pole separation. It remains elusive as how the interaction of CENP-E with PRC1 guides the central spindle elongation and resolution in telophase.
To confirm the interaction of CENP-E with PRC1, we carried out immunoprecipitation experiment using mitotic HeLa cell lysates. As shown in Figure 5D, CENP-E antibody, but not control immunoglobulin G (IgG), pulled down PRC1 protein along with CENP-E (top two panels, lane 3). This indicates that PRC1 is a cognate binding protein of CENP-E in mitosis. To further pinpoint the binding interface between the physical contact of CENP-E–PRC1 interaction, recombinant GST-PRC1, Histidine-CENP-E, GFP-CENP-E, and their deletion mutant proteins were engineered as illustrated in Supplementary Figure S4A and B. Recombinant GST-PRC1 was purified from bacteria and used as affinity matrix to absorb GFP-CENP-E and its deletion mutants from mitotic HeLa cell lysates. After extensive washing, GST-PRC1 and its associated proteins were fractionated on SDS-PAGE followed by western blotting analyses with an anti-GFP antibody. As shown in Supplemental Figure S4C, this pull-down assay showed that GFP-CENP-E C-terminal (GFP-CENP-E1572–2701) possesses the binding activity to the PRC1 N-terminal (GST-PRC11–140). To validate whether CENP-E directly binds to PRC1, we further characterized the PRC1–CENP-E binding interface. Specifically, we employed GST-PRC1 as affinity matrix to isolate purified histidine-tagged CENP-E deletion mutant proteins from bacteria. Recombinant bacterial CENP-E and its deletion mutant proteins were purified and incubated with GST-PRC1 beads. After extensive washing, PRC1 and its associated proteins were fractionated on SDS-PAGE followed by western blotting analyses with an anti-histidine antibody. As shown in Supplementary Figure S4D, CENP-E tail (CENP-E2602–2701) physically binds to the N-terminal PRC1 (GST-PRC11–140). Thus, we conclude that PRC1 binds to CENP-E and the CENP-E–PRC1 interaction in anaphase is essential for central spindle midzone establishment.
Discussion
The functions of epithelial tissue rely on the organized architecture of variety of epithelial cells with uniform polarity. The homeostatic renewal of epithelial cells requires accurate cell division within epithelia. We investigated cell division in gastric organoids to delineate context-dependent physiology and polarity establishment after the exit of cell division. To overcome the sample depth limitation of classical confocal microscope, we adopted Marianas light sheet microscopic imaging system in which dual inverted selective plane illumination was used to visualize live mitosis in organoids. With Marianas light sheet microscope, we were able to image the chromosome segregation and spindle plasticity control in live gastric organoids, and thereby we gained new insights into the mechanisms of chromosome segregation and revealed the temporal function of CENP-E in metaphase–anaphase transition. Together with biochemical characterization and chemical biological interrogation, our study shows that CENP-E interaction with PRC1 orchestrates a critical switch from the lateral association to end-on capture of microtubules (Figure 5E), which provides molecular insight underlying the dynamic remodeling of spindle microtubule into central spindle during metaphase–anaphase transition.
Although CENP-E localizes to midbody and central spindle during metaphase–anaphase, the mechanism of action underlying CENP-E function in central spindle assembly was never addressed despite the fact that an early microinjection experiment suggested a role of CENP-E in anaphase (Yen et al., 1991). Significantly, CENP-E inhibition perturbed the establishment of stable central spindle as syntelin-treated cells exhibited fewer anti-parallel microtubules in anaphase onset. Our findings suggest CENP-E binding to PRC1 contributes to central spindle organization during anaphase onset. The spindle midzone is assembled by incorporating microtubules from the kinetochore microtubules (Eggert et al., 2006; Roostalu and Surrey, 2017). A subset of these spindle microtubules overlaps with anti-parallel orientation while other microtubules such as spindle microtubules terminate at the kinetochore. During metaphase–anaphase transition, CENP-E and PRC1 relocate to the central spindle. Concurrently, stable association of PRC1 with microtubule plus ends promotes cross-linking of the anti-parallel microtubules. Our current study did not address how the interaction of CENP-E–PRC1 is regulated in space and time during metaphase–anaphase transition. Further study will be directed to understanding how microtubule plus ends are tagged by CENP-E–PRC1 as they transit into midzone array and whether post-translational modifications such as phosphorylation and acetylation provide a spatial cue for the selectivity. This can be achieved using FRET-based reporter to illuminate the gradient of mitotic phosphorylation (Chu et al., 2011), methylation (Chu et al., 2012), and acetylation (Bao et al., 2018).
Several recent studies address the cellular dynamics during cell division in organoids. Using intestinal organoids, Vale and colleagues show that the process of cytokinesis in elongated mammalian epithelia allows lineages to intermix and that cellular aspect ratio is a critical modulator of the progeny pattern (McKinley et al., 2018). Using 3D organoid culture systems, Amon and colleagues examine the importance of the tissue environment for chromosome segregation by comparing chromosome segregation fidelity across several cell types in native and non-native contexts (Knouse et al., 2018). They find that physiological tissue architecture promotes the correction of aberrant microtubule-kinetochore attachments during cell division, and this is especially important for maintaining chromosome stability in the polyploid liver. Their findings strengthen the importance of extracellular context in regulating intrinsic cellular processes and the advantage of organoid systems for studying cellular dynamics that naturally function within a tissue. It is worth noting that a combination of small molecule compounds such as these FDA-approved medications with patients-derived and genetically defined organoids will enable us to identify and design individualized precision treatment in clinic oncology.
In sum, our study demonstrates that gastric organoids is a unique model system to delineate the molecular mechanisms underlying context-dependent cell division control in 3D tissue environment. Modeling mitosis in organoids using chemical probes led us to uncover the functional regulation of CENP-E in metaphase–anaphase transition. Finally, the spatiotemporal regulation of CENP-E–PRC1 interaction may provide an insight into molecular delineation of dynamic transition from spindle microtubule into central spindle and cell fate decision control. Future work will aim to identify the spatiotemporal cues within the mitotic spindle to regulate CENP-E–PRC1 interaction for accurate anaphase midzone formation.
Materials and methods
Organoids culture
A detailed protocol for gastric organoids culture was documented (Barker et al., 2010; Bartfeld et al., 2015). Briefly, glands were extracted from 1 cm2 of human gastric mucosa using ethylenediaminetetraacetic acid (EDTA) in cold chelation buffer, seeded in Matrigel (BD Biosciences), and overlaid with medium containing advanced DMEM/F12 supplemented with penicillin/streptomycin, 10 mM HEPES, GlutaMAX, 1 × B27 (all from Invitrogen), and 1 mM N-acetylcysteine (Sigma-Aldrich). Growth factors were added to the basal medium. The final human stomach culture medium contained the following essential components: 50 ng/ml EGF (Invitrogen), 10% noggin-conditioned medium, 10% R-spondin1-conditioned medium, 50% Wnt-conditioned medium, 200 ng/ml FGF10 (Peprotech), 1 nM gastrin (Tocris), and 2 μM TGF-β1 (A-83-01; Tocris). The facultative component was 10 mM nicotinamide (Sigma-Aldrich). After seeding, RHO kinase inhibitor Y-27632 (10 μM; Sigma-Aldrich) was added. In general, the passages 2–8 organoids from five different preparations were used in our experiments without noticed difference in morphology and acid secretory activity in response to histamine stimulation. The generation of human gastric organoids in this study was approved by the Ethics Committee of the Dongzhimen Hospital of the Beijing University of Chinese Medicine, and written informed consent was obtained from all patients.
Cell culture, synchronization, and transfection
HeLa cells and HEK293T cells (American Type Culture Collection) were cultured in DMEM (Invitrogen) with 10% (v/v) FBS (Hyclone) and penicillin–streptomcyin (100 units/ml and 100 μg/ml, respectively; Invitrogen) at 37°C in a humidified atmosphere with 8% CO2. For cell cycle synchronization, HeLa cells were first blocked in G1/S with 2.5 mM thymidine (Sigma) for 16 h and then released in fresh culture medium for 8 h to enrich mitotic cells (Akram et al., 2018). For some experiments, cells were exposed to 10 μM MG132 and 1 μM syntelin or DMSO for another 1 h before fixation. Plasmid transfections were performed with Lipofectamine 3000 (Invitrogen) according to the manufacturer’s instructions.
Antibodies and chemical inhibitors
Anti-α-tubulin antibody (mouse, FITC-DM1A, F2168; Sigma-Aldrich), anti-CENP-A antibody (mouse, ab13939; Abcam), anti-MKLP1 antibody (rabbit, ab204478; Abcam), and Rhodamine-conjugated phalloidin (R415; Invitrogen) were used for immunofluorescence. The appropriate secondary antibodies were purchased from Jackson ImmunoResearch Laboratories. Antibodies used for western blots were anti-α-tubulin (mouse, DM1A, T9017; Sigma-Aldrich) and anti-BubR1 (mouse, 612503; BD Biosciences). Anti-CENP-E antibody (HpX) was generated as previously described (Yao et al., 1997). Anti-PRC1 antibody used in immunofluorescence and western blot was generated as described (Fu et al., 2007).
Nocodazole (100 ng/ml), monastrol (50 μM), MG132 (10 μM), and reversine (300 nM) were from Sigma. GSK923295 (100 nM) and BI2536 (100 nM) were from Selleckchem. Syntelin (1 μM) was synthesized as described before (Ding et al., 2010). The protease inhibitor cocktail was from Sigma.
Recombinant protein expression and purification
Recombinant human His-CENP-E truncation plasmids were transformed into Escherichia coli strain Rosetta (DE3) and protein expression was induced with 0.5 mM IPTG at 16°C for 10 h. Bacteria expressing His-tagged CENP-E proteins were suspended and lysed in lysis buffer (50 mM NaH2PO4, pH 8.0; 300 mM NaCl; 10 mM imidazole) with 1 μg/ml PMSF. His-tagged proteins were bound to Ni-NTA resin (Qiagen) and eluted with elution buffer (50 mM NaH2PO4, pH 8.0; 300 mM NaCl; 200 mM imidazole) for corresponding experiments. All purification procedures were performed at 4°C, and protease inhibitor cocktail was added to prevent protein degradation.
GST-PRC1 truncation plasmids were transformed into E. coli strain Rosetta (DE3), and protein expression was induced with 0.5 mM IPTG at 16°C. Bacteria expressing GST-PRC1 were suspended and lysed by sonication in phosphate buffered saline (PBS) buffer supplemented with 1% Triton X-100. The preparation was incubated with glutathione-Sepharose 4B (GE Healthcare) for 1.5 h at 4°C. The resin was washed three times, and GST-H3 protein was eluted with 10 mM glutathione.
Immunoprecipitation
For immunoprecipitation, synchronized HeLa cells were lysed in IP buffer (50 mM Tris-HCl, pH 8.0; 120 mM NaCl; 0.2% NP-40) supplemented with protease inhibitor cocktail. After pre-clearing with protein A/G resin (ThermoFisher Scientific), the lysate was incubated with CENP-E antibody at 4°C for 24 h with gentle rotation. Protein A/G resin was then added into the cell lysates to incubate for another 6 h followed by spun down and washed five times with lysis buffer. The immunoprecipitates were resolved by SDS-PAGE and immunoblotted with the indicated antibodies.
For pull-down assays, GST-PRC1-bound sepharose beads were incubated with HEK293T cell lysates containing ectopically expressed GFP-tagged CENP-E or with purified His-tagged CENP-E from bacteria in PBS containing 0.2% Triton X-100 at 4°C for 4 h. The binding fraction was washed with PBS for three times and analyzed by Coomassie Brilliant Blue stained SDS-PAGE gel.
Syntelin affinity pull-down assay
Mitotic HeLa cell lysates were incubated with biotin-syntelin (5 μM) or DMSO for 2 h. Then the cells were lysed in lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.1% (v/v) Triton X-100, 5 mM EDTA and proteinase inhibitors) and centrifuged at 16000 g for 20 min at 4°C. The supernatant was incubated with Avidin Agarose (ThermoFisher Scientific) for 2 h. The beads were washed three times with lysis buffer and then resuspended in 50 μl lysis buffer with 10 μM syntelin to compete with biotin-syntelin. After 30-min incubation for competition, supernatant was removed and SDS-PAGE sample buffer was added into the beads. The samples were resolved by SDS-PAGE and immunoblotted with indicated antibodies.
Immunofluorescence and light sheet microscope
HeLa cells growing on coverslips after transfection or drug treatment were extracted by pre-warmed PHEM (60 mM PIPES, 25 mM HEPES, pH 6.9, 10 mM EGTA, 2 mM MgCl2, and 4 M Glycerol), followed by 1 min of permeabilization with PHEM containing 0.1% Triton X-100. The extracted cells were then fixed with 3.7% paraformaldehyde in PHEM for 5 min. After being washed three times with PBST (0.05% Tween-20 in PBS), cells were blocked with 1% bovine serum albumin (BSA; Sigma) in PBST for 1 h, then incubated with primary antibodies for 1 h and secondary antibodies for 1 h at room temperature. DNA was stained with DAPI (Sigma). Images of 2D cell culture were acquired every 0.25 μm at z axis to generate 3D image stacks using an Olympus 60×/1.42 Plan APO N objective on an Olympus IX71 microscope (Applied Precision DeltaVision). The 3D image stacks were deconvolved and projected with SoftWorx (Applied Precision) and mounted in figures with Photoshop and Illustrator (Adobe). Fluorescence was qualified with ImageJ software (NIH).
For 3D organoids imaging, gastric organoids were fixed with 4% paraformaldehyde in PBS for 2 h. After being washed three times with PBST (0.05% Tween-20 in PBS), organoids were treated with 0.2% Triton X-100 in PBS for 2 h and blocked with 1% BSA (Sigma) in PBST for 1 h, then incubated with primary antibodies for 1 h (DM1A anti-α-tubulin antibody; Sigma) and secondary antibodies for 1 h at room temperature. DNA was stained with DAPI (Sigma). Fixed and stained samples were acquired every 0.5 μm at Z axis to generate 3D image stacks using a 3i Marianas LightSheet fluorescence microscope (Intelligent Imaging Innovations, Inc.), fitted with 10× illumination objectives and an achromatic 40× detection objective. The laser wavelengths used were 405 nm, 488 nm, and 561 nm and the laser intensity was kept to a minimum. Fluorescent beads (ThermoFisher Scientific) were used as fiduciary markers and calibration.
Data analysis was performed using the 3i analysis software slidebook6. Three-dimensional visualizations of the data were generated using the 3D Volume View of Slidebook6. The positional data and numerical features were generated using MultiD channel View of slidebook6.
Live cell imaging
Time-lapse imaging of cultured cells was accomplished with a 60× oil-immersion 1.42 numerical aperture objective lens on an Olympus IX71 microscope as previously described (Xia et al., 2012). Images were recorded at 37°C in Lab-Tek Chambered 1.0 Borosilicate Coverglass system (Nunc) or MatTek glass-bottom microwell dishes (MatTek) in CO2-independent medium (Gibco) containing 10% (v/v) FBS, 100 units/ml penicillin, and 100 μg/ml streptomycin. For time-lapse microscopy of chromosome segregation in mitotic cells, images were taken with 500-ms exposures for mCherry-tubulin and 100-ms exposures for GFP-PRC1 and GFP-H2B every 2–3 min from 24 h after transfection. The frames were then projected with SoftWORX software (Applied Precision) and mounted by Photoshop and Illustrator (Adobe).
Electron microscopy
To understand the ultrastructure of central spindle in CENP-E-inhibited cells, we carried out immuno-electron microscopic analyses of HeLa cells treated with syntelin at metaphase using an established protocol (Yao et al., 1997). Specifically, HeLa cells were treated as illustrated in Figure 2A followed by fixation with 1% glutaraldehyde (Tousimis) in PBS. After three washes in PBS, fixed cells were then postfixed in 2% osmium tetroxide (Electron Microscopy Sciences), dehydrated in a graded alcohol series followed by 100% acetone, and embedded in Epoxy (Ernest F. Fullam, Inc.). The cells were detached from coverslips using hydrofluoric acid, and the designated areas were excised and glued to blocks. Thin serial sections (silver–gold) were then cut, placed on copper grids, and stained with uranyl acetate and lead citrate. The immuno-gold labeling was monitored by 10-nm gold conjugated goat anti-rabbit IgG. Immunostained grids were stained with uranyl acetate before examination. The sections were examined by a JEOL 1200 EM.
Statistical analysis
All statistics were described in the figure legends. Two-sided unpaired Student’s t-test was applied for experimental comparisons, using GraphPad Prism.
Supplementary Material
Acknowledgements
We are grateful to Prof. Yunyu Shi for the support, Drs Ye-Guang Chen and Richard Peek for their input in organoids modeling. We thank all the members of our laboratories for insightful discussion and suggestions.
Funding
This work was supported in part by the National Natural Science Foundation of China (31430054, 31320103904, 31621002, 31671405, 31601097, 91854203, 91753000, and 91853115), ‘Strategic Priority Research Program’ of the Chinese Academy of Sciences (XDB19000000), the National Key Research and Development Program of China (2017YFA0503600 and 2016YFA-0100500), MOE Innovative Team project (IRT_17R102), and the US National Institutes of Health (CA164133, DK56292, and DK115812).
Conflict of interest: none declared.
References
- Adams, G., Zhou, J., Wang, W., et al. (2016). Microtubule plus-end tracking protein TIP150 interacts with cortactin to steer directional cell migration. J. Biol. Chem. 291, 20692–20706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhmanova, A., and Steinmetz, M.O. (2008). Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nat. Rev. Mol. Cell Biol. 9, 309–322. [DOI] [PubMed] [Google Scholar]
- Akram, S., Yang, F., Li, J., et al. (2018). LRIF1 interacts with HP1alpha to coordinate accurate chromosome segregation during mitosis. J. Mol. Cell Biol. 10, 527–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhoum, S.F., Ngo, B., Laughney, A.M., et al. (2018). Chromosomal instability drives metastasis through a cytosolic DNA response. Nature 553, 467–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao, X., Liu, H., Liu, X., et al. (2018). Mitosis-specific acetylation tunes Ran effector binding for chromosome segregation. J. Mol. Cell Biol. 10, 18–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker, N., Huch, M., Kujala, P., et al. (2010). Lgr5+ve stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell 6, 25–36. [DOI] [PubMed] [Google Scholar]
- Bartfeld, S., Bayram, T., van de Wetering, M., et al. (2015). In vitro expansion of human gastric epithelial stem cells and their responses to bacterial infection. Gastroenterology 148, 126–136.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjerknes, M., and Cheng, H. (2002). Multipotential stem cells in adult mouse gastric epithelium. Am. J. Physiol. Gastrointest. Liver Physiol. 283, G767–G777. [DOI] [PubMed] [Google Scholar]
- Chu, L., Zhu, T., Liu, X., et al. (2012). SUV39H1 orchestrates temporal dynamics of centromeric methylation essential for faithful chromosome segregation in mitosis. J. Mol. Cell Biol. 4, 331–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu, Y., Yao, P.Y., Wang, W., et al. (2011). Aurora B kinase activation requires survivin priming phosphorylation by PLK1. J. Mol. Cell Biol. 3, 260–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland, D.W., Mao, Y., and Sullivan, K.F. (2003). Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell 112, 407–421. [DOI] [PubMed] [Google Scholar]
- Ding, M., Jiang, J., Yang, F., et al. (2019). Holliday junction recognition protein interacts with and specifies the centromeric assembly of CENP-T. J. Biol. Chem. 294, 968–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, X., Yan, F., Yao, P., et al. (2010). Probing CENP-E function in chromosome dynamics using small molecule inhibitor syntelin. Cell Res. 20, 1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou, Z., Liu, X., Wang, W., et al. (2015). Dynamic localization of Mps1 kinase to kinetochores is essential for accurate spindle microtubule attachment. Proc. Natl Acad. Sci. USA 112, E4546–E4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggert, U.S., Mitchison, T.J., and Field, C.M. (2006). Animal cytokinesis: from parts list to mechanisms. Annu. Rev. Biochem. 75, 543–566. [DOI] [PubMed] [Google Scholar]
- Eicher, A.K., Berns, H.M., and Wells, J.M. (2018). Translating developmental principles to generate human gastric organoids. Cell. Mol. Gastroenterol. Hepatol. 5, 353–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forte, J.G., and Yao, X. (1996). The membrane-recruitment-and-recycling hypothesis of gastric HCl secretion. Trends Cell Biol. 6, 45–48. [DOI] [PubMed] [Google Scholar]
- Fu, C., Yan, F., Wu, F., et al. (2007). Mitotic phosphorylation of PRC1 at Thr470 is required for PRC1 oligomerization and proper central spindle organization. Cell Res. 17, 449–457. [DOI] [PubMed] [Google Scholar]
- Gudimchuk, N., Vitre, B., Kim, Y., et al. (2013). Kinetochore kinesin CENP-E is a processive bi-directional tracker of dynamic microtubule tips. Nat. Cell Biol. 15, 1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Y., Lin, L., Liu, X., et al. (2019). BubR1 phosphorylates CENP-E as a switch enabling the transition from lateral association to end-on capture of spindle microtubules. Cell Res. 29, 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor, T.M., Lampson, M.A., Hergert, P., et al. (2006). Chromosomes can congress to the metaphase plate before biorientation. Science 311, 388–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, Y., Holland, A.J., Lan, W., et al. (2010). Aurora kinases and protein phosphatase 1 mediate chromosome congression through regulation of CENP-E. Cell 142, 444–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knouse, K.A., Lopez, K.E., Bachofner, M., et al. (2018). Chromosome segregation fidelity in epithelia requires tissue architecture. Cell 175, 200–211.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J., Wang, Z., Jiang, K., et al. (2009). PRC1 cooperates with CLASP1 to organize central spindle plasticity in mitosis. J. Biol. Chem. 284, 23059–23071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer, T.U., Kapoor, T.M., Haggarty, S.J., et al. (1999). Small molecule inhibitor of mitotic spindle bipolarity identified in a phenotype-based screen. Science 286, 971–974. [DOI] [PubMed] [Google Scholar]
- McCracken, K.W., Catá, E.M., Crawford, C.M., et al. (2014). Modelling human development and disease in pluripotent stem-cell-derived gastric organoids. Nature 516, 400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald, S.A., Greaves, L.C., Gutierrez-Gonzalez, L., et al. (2008). Mechanisms of field cancerization in the human stomach: the expansion and spread of mutated gastric stem cells. Gastroenterology 134, 500–510. [DOI] [PubMed] [Google Scholar]
- McKinley, K.L., Stuurman, N., Royer, L.A., et al. (2018). Cellular aspect ratio and cell division mechanics underlie the patterning of cell progeny in diverse mammalian epithelia. eLife 7, e36739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo, F., Zhuang, X., Liu, X., et al. (2016). Acetylation of Aurora B by TIP60 ensures accurate chromosomal segregation. Nat. Chem. Biol. 12, 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roostalu, J., and Surrey, T. (2017). Microtubule nucleation: beyond the template. Nat. Rev. Mol. Cell Biol. 18, 702–710. [DOI] [PubMed] [Google Scholar]
- Steegmaier, M., Hoffmann, M., Baum, A., et al. (2007). BI 2536, a potent and selective inhibitor of polo-like kinase 1, inhibits tumor growth in vivo. Curr. Biol. 17, 316–322. [DOI] [PubMed] [Google Scholar]
- Subramanian, R., Ti, S.-C., Tan, L., et al. (2013). Marking and measuring single microtubules by PRC1 and kinesin-4. Cell 154, 377–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward, T., Wang, M., Liu, X., et al. (2013). Regulation of a dynamic interaction between two microtubule-binding proteins, EB1 and TIP150, by the mitotic factor PCAF orchestrates kinetochore microtubule plasticity and chromosome stability during mitosis. J. Biol. Chem. 288, 15771–15785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood, K.W., Lad, L., Luo, L., et al. (2010). Antitumor activity of an allosteric inhibitor of centromere-associated protein-E. Proc. Natl Acad. Sci. USA 107, 5839–5844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, P., Wang, Z., Liu, X., et al. (2012). EB1 acetylation by P300/CBP-associated factor (PCAF) ensures accurate kinetochore-microtubule interactions in mitosis. Proc. Natl Acad. Sci. USA 109, 16564–16569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, P., Zhou, J., Song, X., et al. (2014). Aurora a orchestrates entosis by regulating a dynamic MCAK–TIP150 interaction. J. Mol. Cell Biol. 6, 240–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, X., Abrieu, A., Zheng, Y., et al. (2000). CENP-E forms a link between attachment of spindle microtubules to kinetochores and the mitotic checkpoint. Nat. Cell Biol. 2, 484–491. [DOI] [PubMed] [Google Scholar]
- Yao, X., Anderson, K.L., and Cleveland, D.W. (1997). The microtubule-dependent motor centromere-associated protein E (CENP-E) is an integral component of kinetochore corona fibers that link centromeres to spindle microtubules. J. Cell Biol. 139, 435–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, X., and Forte, J.G. (2003). Cell biology of acid secretion by the parietal cell. Annu. Rev. Physiol. 65, 103–131. [DOI] [PubMed] [Google Scholar]
- Yao, X., and Smolka, A.J. (2019). Gastric parietal cell physiology and helicobacter pylori-induced disease. Gastroenterology 156, 2158–2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen, T.J., Compton, D., Wise, D., et al. (1991). CENP-E, a novel human centromere-associated protein required for progression from metaphase to anaphase. EMBO J. 10, 1245–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.





