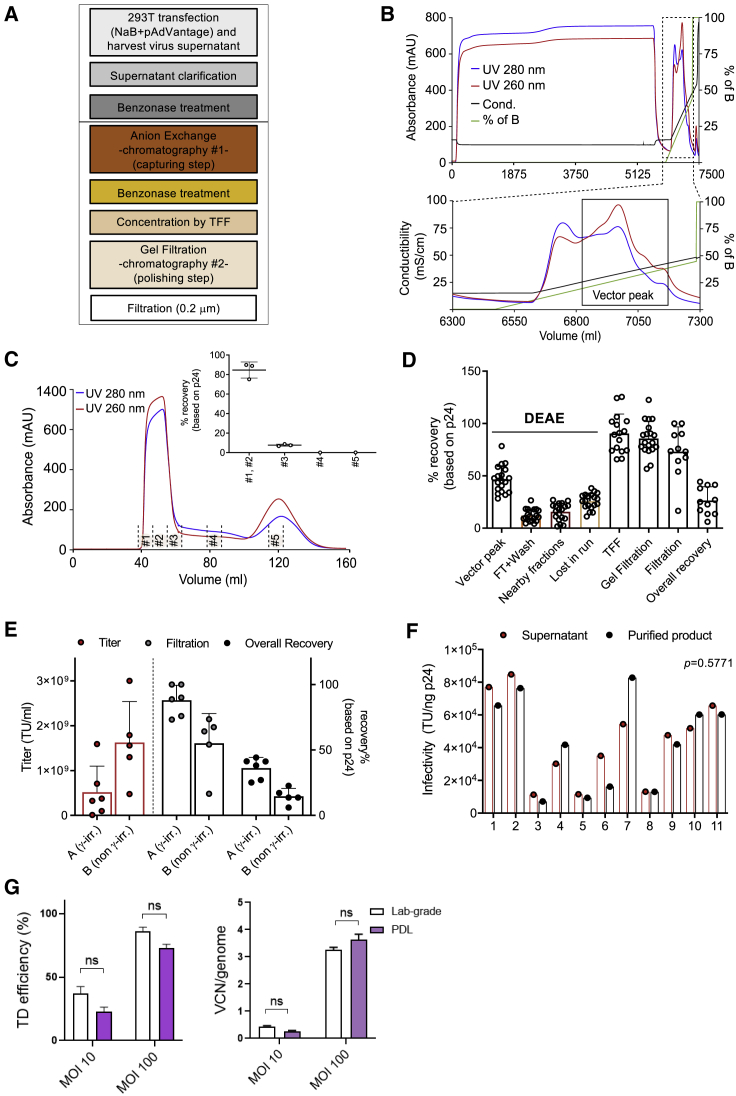
Figure 2.
Optimization of LV Purification
(A) Overview of the purification workflow. (B) Linear gradient elution profile obtained after loading of the LV-containing supernatant on a column packed with the DEAE anion exchanger. Zoomed box shows the eluted peaks (black line, conductivity in mS/cm; green line, percentage of buffer B [PBS + 1 M NaCl]; blue and red lines, absorbance at 280 and 260 nm (mAU), respectively; on the x axis, changes in volume during elution). (C) Chromatographic profile of LV after gel filtration (GF) chromatography (blue and red lines, absorbance at 280 and 260 nm [mAU], respectively). In the box, the recovery after elution is shown as percentage of physical particles. Data are plotted as mean with SD (n ≥ 3) or as mean with range (n < 3). #1–#5 indicate the fractions collected during the GF run. (D) Physical particles calculated as percentage of recovery for different steps of the downstream process. Data are plotted as mean with SD (n = 19 for DEAE and GF, n = 16 for TFF, and n = 11 for filtration and overall recovery step). (E) Infectious titer (TU/mL, plotted on left y axis) and physical particles calculated as percentage of recovery (plotted on right x axis). In group A, only one vector was produced with non-γ-irradiated serum. Data are plotted as mean with SD (total n = 11). (F) Infectivity of the particles of the LV-containing supernatant and purified LV. Significance was assessed with a Wilcoxon matched-pairs signed-rank test. (G) Laboratory-grade (lab-grade) and purified (PDL) LV transduction efficiencies in mobilized peripheral blood (mPB)-CD34+ HSPCs measured by flow cytometry 5 days post-transduction (left) and by ddPCR 14 days post-transduction (right) (mean ± SEM; lab grade n = 3; PDL n = 3).