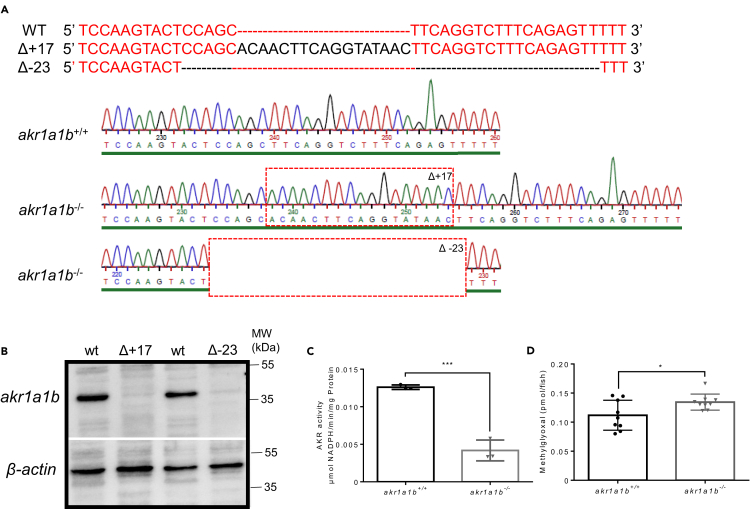
Figure 2.
Generation and Validation of akr1a1b-/- Zebrafish Mutants
(A) CRISPR-Cas9 technology was used to establish akr1a1b knockout zebrafish. Schematic depiction of wild-type akr1a1b target sequence and two identified frameshift mutations and their corresponding chromatograms including a 17-bp insertion (Δ+17) in the Tg(wt1b:EGFP) reporter line and a 23-bp deletion (Δ-23) in the Tg(fli1:EGFP) reporter line. Red dashed boxes indicate start of genomic alterations.
(B) Western blot for Akr1a1b expression in zebrafish livers showed absence of Akr1a1b protein in the 17-bp insertion (Δ+17) and in the 23-bp deletion mutant (Δ-23), respectively, which validates the akr1a1b knockout zebrafish model. Beta-actin served as loading control.
(C) Δ+17 akr1a1b-/- zebrafish larvae at 96 hpf showed a strong descend of Akr enzyme activity (n = 3 clutches with 50 larvae, mean ± SD).
(D) Δ+17/Δ-23 akr1a1b-/- larvae at 96 hpf have increased MG concentrations (n = 9 clutches with 50 larvae).
∗p < 0.05, ∗∗∗p < 0.001, p value was calculated by t test. See also Figure S2.