Figure 6.
TBsRNA-33 Is an Anti-Sense Regulator of T. brucei Pif1 mRNA
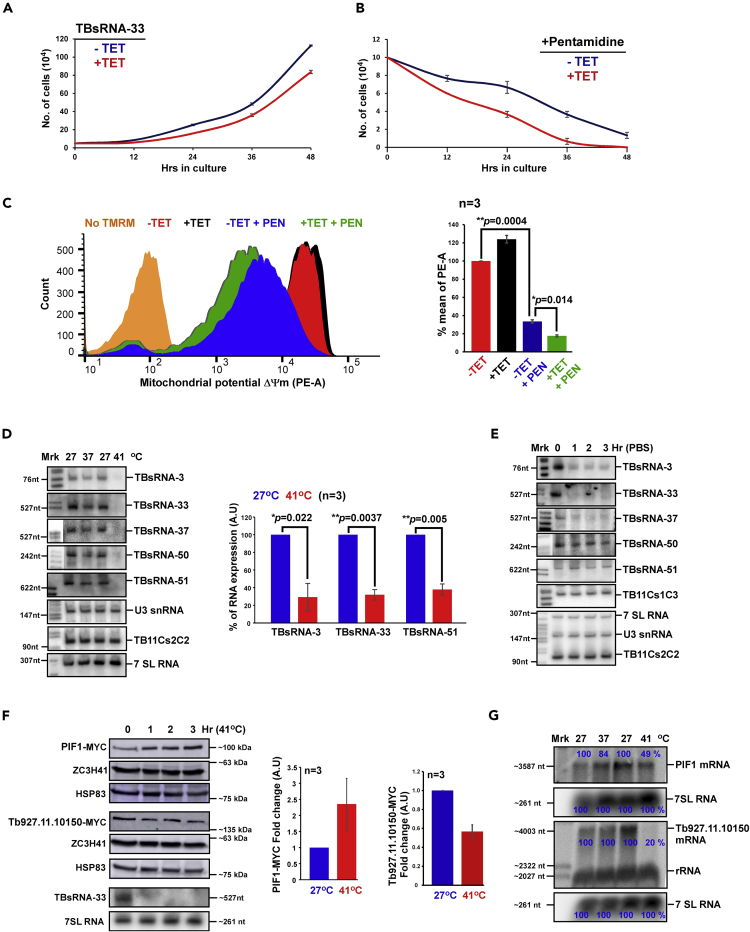
(A) Growth of cells upon TBsRNA-33 silencing. Uninduced cells carrying the silencing construct (-TET) were compared with cells induced for silencing (+TET) at 27°C. Data are presented as mean ± S.E.M. Experiments were done in triplicate (n = 3).
(B) Growth of cells upon TBsRNA-33 silencing and pentamidine treatment. Uninduced cells carrying the silencing construct (-TET) were compared with cells induced for silencing (+TET) upon treatment with 20μM pentamidine. Data are presented as mean ± S.E.M. Experiments were done in triplicate (n = 3).
(C). Silencing of TBsRNA-33 induces loss of mitochondrial membrane potential upon pentamidine treatment. Uninduced cells (-TET) and cells induced for silencing (+TET) upon treatment with 20μM pentamidine were harvested and treated with 150 nM TMRM in PBS with 0.2% glucose. The samples were incubated in the dark for 15 min at 27°C and then analyzed by FACS in PE channel. The cell count of cells with reduced ΔΨm is plotted comparing the cells before and after two days of silencing. Data are presented as mean ± S.E.M (left). Experiments were done in triplicate (n = 3). Student's t-test was performed, and the p value is indicated.
(D) TBsRNAs are reduced upon heat-shock at 41°C. Total RNA (20 μg) from PCF cells subjected to heat-shock for 2 hr at the temperature indicated was separated on a 6% or 10% denaturing polyacrylamide gel and detected by Northern analysis with 32P-labeled anti-sense RNA probes to TBsRNA (left). The quantitative data derived from Northern analysis is presented as mean ± S.E.M (right). Experiments were done in triplicate (n = 3). Student's t-test was performed, and the p value is indicated. 7SL RNA was used as a loading control. 32P-labeled pBR322 DNA MspI digest was used as a size marker.
(E) TBsRNAs are regulated upon starvation. Total RNA (20 μg) from PCF cells upon starvation in 1X PBS for the indicated time points was separated on 6% or 10% denaturing polyacrylamide gels and detected by Northern analysis.
(F) Western analysis of PIF1 protein following heat-shock. Total cell lysates were prepared from an equal number of cells before and after heat-shock and was subjected to western analysis using the indicated antibodies (left). The dilutions used for the antibodies are: (A) MYC (1:10,000), HSP83 (1:10,000), MTAP (1:10,000), and ZCH341 (1:10,000). The RNA derived from the same cells used for Western blot was subjected to Northern analysis. The data for PIF1-MYC is presented as mean ± S.E.M (right). Experiments were done in triplicate (n = 3). The expression level of HSP83 was used as a loading control.
(G) Northern analysis of Pif1 mRNA following heat-shock. Total RNA (20 μg) was prepared under the conditions indicated, separated on a 1.2% agarose/formaldehyde gel and subjected to Northern analysis using RNA probes, as indicated. All densitometry data used in this figure was calculated by ImageJ (https://imagej.nih.gov/ij/) software