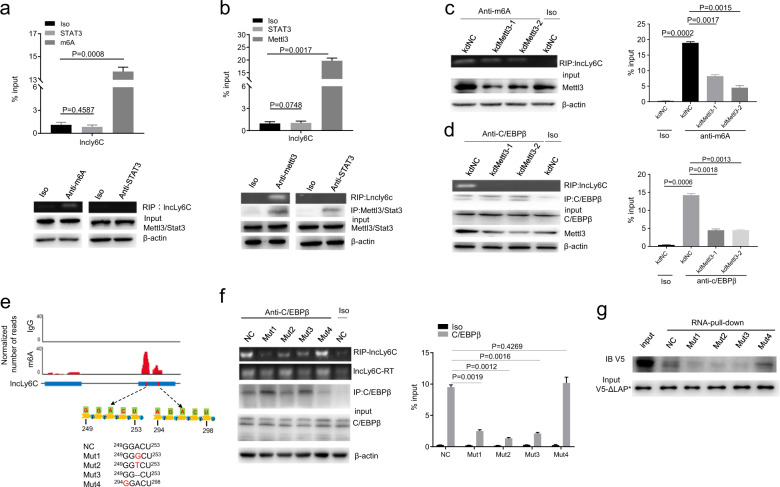
Fig. 5. Binding of lncLy6C to C/EBPβ depends on m6A methylation.
a RIP analyses in BMDMs after exposed to butyrate. RIP was performed using anti-m6A antibody and then PCR for lncLy6C. Iso, isotypic antibody; anti-STAT3 (STAT3), negative control. % input was compared. b RIP analyses in BMDMs after exposed to butyrate (200 μM). RIP was performed using anti-Mettl3 antibody and then PCR for lncLy6C. Iso, isotypic antibody; anti-STAT3 (STAT3), negative control. % input was compared. c RIP analyses in mettl3 siRNA transfected BMDMs. RIP was performed using anti-m6A antibody and then PCR for lncLy6C. kdNC, siRNA control; KdMettl3-1 and KdMettl3-2, different mettl3 siRNA; Iso, isotypic antibody. % input (lower) was compared. d RIP analyses of mettl3 siRNA transfected BMDMs. RIP was performed using anti-C/EBPβ antibody and then PCR for lncLy6C. kdNC, siRNA control; KdMettl3-1 and KdMettl3-2, different mettl3 siRNAs; Iso, isotypic antibody. % input (lower) was compared. e m6A RIP-seq of lncLy6C and sequence logo of lncLy6C with m6A methylation motifs. m6A peaks are enriched in the lncLy6C gene. The iCLIP truncations track shows the positions of iCLIP cDNA truncations at lncRNA with peak height corresponding to cDNA counts. Schematic of the corresponding regions of CEBP/β binding motifs in lncRNA (low) and the mutant base in motif portions are labeled in red. f RIP-PCR analysis in HEK293T cells cotransfected with C/EBPβ expression vector and different lncRNA mutants. RIP was performed using anti-CEBP/β and then PCR for lncRNA. % input of lncLy6C was analyzed (lower). NC, full length lncLy6C. g RNA pull down using biotinylated lncLy6C (NC) or lncLy6C mutants in V5-tagged C/EBPβ LAP* transfected HEK293T cells. One-way ANOVA and Bonferroni’s multiple comparison test in b. Two-sided Student’s t test in c. ANOVA and Bonferroni’s multiple comparison test in a–d and f.