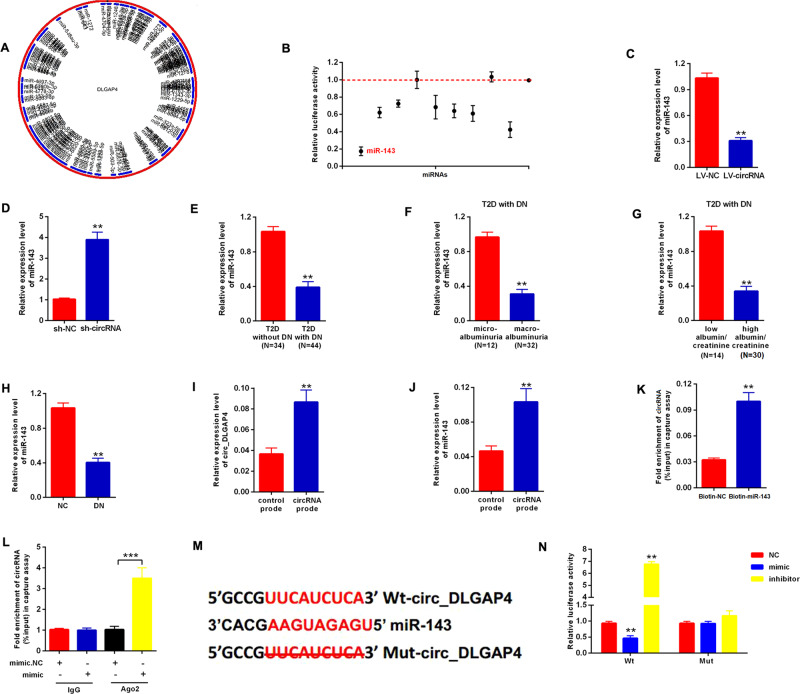
Fig. 3. miR-143 was the direct target of circ_DLGAP4.
A The schematic model indicated the putative binding sites of predicted miRNAs on circ_DLGAP4 (http://www.bioinf.com.cn/). B Luciferase activity of circ_DLGAP4 in MCs transfected with miRNA mimics with putative binding to the circ_DLGAP4 sequence. C, D The expression level of miR-143 in MCs transfected with LV-circ_DLGAP4 or sh-circ_DLGAP4. E The expression level of miR-143 in T2D patients without DKD (n = 34) or with DKD (n = 44). F Expression levels of miR-143 was detected in DKD with microalbuminuria (n = 12) or with macroalbuminuria (n = 32). G The expression level of miR-143 were detected in DKD with low eGFR (n = 14) or high eGFR (n = 30). H The expression level of miR-143 was decreased in rat DKD model. I circ_DLGAP4 in cell lysis was pulled down and enriched with circ_DLGAP4 specific probe in MCs. J miR-143 was pulled down and enriched with circ_DLGAP4-specific probe in MCs. K Biotin-coupled miR-143 captured a fold change of circ_DLGAP4 in the complex as compared with biotin-coupled NC in biotin-coupled miRNA capture. L RIP was performed using Ago2 antibody in MCs cells transfected with miR-143 mimic or mimic NC. The enrichment of circ_DLGAP4 was detected. M The direct binding sites between circ_DLGAP4 and miR-143 were presented. N Luciferase reporter activity of circ_DLGAP4 in MCs cells co-transfected with miR-143 mimic, inhibitor, or NC. Three independent experiments were conducted. Error bars stand for the mean ± SD of at least triplicate experiments; **P < 0.01, ***P < 0.001.