Abstract
SMARCA4 and EZH2 are two functional key players of their respective antagonizing chromatin remodeling complexes SWI/SNF and PRC2. EZH2 inhibitory drugs may abrogate pro-oncogenic features of PRC2 and turn the balance to cell differentiation via SWI/SNF activity in cancers. SMARCA4 and EZH2 expression was assessed by RT-PCR in 238 epithelial ovarian cancers (OCs) and put in relation to clinico-pathological parameters and patients’ outcome. Optimal thresholds for high and low expression of both variables were calculated by the Youden’s index based on receiver operating characteristic (ROC) curves. High SMARCA4 mRNA expression was independently associated with favorable progression-free survival (PFS) (P = 0.03) and overall survival (OS) (P = 0.018). As Youden’s threshold determination for EZH2 yielded a S-shaped ROC-curve, two cut-off points (29th and 94th percentile) predicting opposite features were defined. Whereas EZH2 mRNA levels beyond the 29th percentile independently predicted poor PFS (P = 0.034), Cox-regression in EZH2 transcripts above the 94th percentile revealed a conversion from unfavorable to favorable PFS and OS (P = 0.009 and P = 0.032, respectively). High SMARCA4 expression associates with improved survival, whereas moderate/high EZH2 expression predicts poor outcome, which converts to favorable survival in ultra-high expressing OCs. This small OC subgroup could be characterized by REV7-abrogated platinum hypersensitivity but concomitant PARP-inhibitor resistance.
Subject terms: Chromatin remodelling, Ovarian cancer, Ovarian cancer
Introduction
Ovarian cancer (OC) is the leading cause of death in women with gynecological malignancies in the developed world1, 2. Although during the last decade implementation of a number of new treatment approaches against OC led to an improvement of prognosis, the latter still remains disastrous3.
The accessibility of DNA to regulatory transcription machinery proteins is regulated by antagonizing classes of enzymes which are found in two large ATP-dependent protein complexes, the so-called chromatin remodeling complexes, namely the switch/sucrose non-fermentable (SWI/SNF) complex and the covalent histone-modifying complexes, like the polycomb repressive complex (PRC2). These complexes regulate the structure of chromatin by either activating or repressing functions and thereby are crucial for genomic transcription and gene expression4,5.
The SWI/SNF complex uses the energy from ATP hydrolysis to weaken interactions between DNA and histones and thereby regulates the access to genes for the mechanisms of DNA transcription, replication and repair6,7. Members of the SWI/SNF complex are recurrently mutated and inactivated in about 20% of human cancers8 and the tumor suppressive function of SWI/SNF is well documented. The human SWI/SNF complex contains only one of the two mutually exclusive catalytic subunits with ATPase activity—either SMARCA2 (BRM) or SMARCA4 (BRG1)—and several other subunits, including ARID1A and ARID1B, which are mutated in a variety of cancers6,7. Mutations of ARID1A can be found in ovarian clear-cell and endometrioid carcinomas9 as well as in high-grade endometrial carcinomas10. ARID1B can be used as a prognostic and predictive biomarker in breast cancer11. PBRM1, another gene of SWI/SNF complex is frequently mutated in renal cancer12. Malignant rhabdoid tumors often harbor mutations of SMARCB113.
The expression of SMARCA4, which is one of the main topics in this paper, is absent in around 10% of human primary non-small-cell lung cancers (NSCLC) and its loss is associated with poor patient survival14. Interestingly, inactivating biallelic mutations of SMARCA4 gene can be found in nearly all small cell carcinoma of the ovary of the hypercalcemic type (SCCOHT), which was first described in 1979 by R. Scully15–18. SCCOHT is a very rare but aggressive form of ovarian cancer diagnosed especially in young women under the age of 40 years. It is characterized by elevated paraneoplastic serum calcium levels and often occurs within families15,19. Recurrence is rapid and approximately two thirds of patients with advanced disease die within 2 years after diagnosis20.
In contrary, proteins of the PRC2 complex play an important role in oncogenic transformation21. Its catalytic subunit EZH2 (Enhancer of zeste homolog 2) trimethylates lysine 27 of histone H3 (H3K27me) and thereby promotes transcriptional silencing. EZH2 is highly expressed in various tumors, such as breast cancer, prostate cancer, endometrial cancer and melanoma, where it often correlates with advanced tumor stages and poor prognosis22–24. Recently E2F1 was found to directly regulate transcription of EZH2 and SUZ12, another component of the PRC2 complex. Furthermore, E2F1-EZH2-SUZ12 signature predicts aggressiveness and impaired prognosis in bladder cancer25. In epithelial OCs EZH2 is also often overexpressed and thereby promotes proliferation and invasion of tumor cells26.
Inhibition of EZH2 leads to reduced tumor formation and growth. Therefore, EZH2 inhibitors, such as tazemetostat, could play a potential role in the treatment of many cancers27,28. The Food and Drug Administration granted recently accelerated approval to tazemetostat for metastatic or locally advanced epithelioid sarcoma not eligible for complete resection.
In SCCOHT it was shown that EZH2 inhibitors induced cell cycle arrest, apoptosis and cell differentiation in tumor cells as well as suppression of tumor growth and improved survival in murine models18. Furthermore, in high-grade serous cancers (HGSOC) another mechanism involving EZH2 was recently reported in CARM1 overexpressing cancers. Herein authors showed that EZH2 inhibitors sensitizes this OC subtype to PARP inhibitors despite its proficiency in homologous recombination DNA repair29.
The high prevalence of SWI/SNF mutations in various human malignancies, the occurrence of SMARCA4 as the driver mutation in SCCOHT and the potential therapeutic strategy of EZH2 inhibitors led us to explore the role of SMARCA4 and EZH2 in epithelial ovarian cancers. Therefore, we determined SMARCA4 and EZH2 expression on the transcriptome level in 238 fresh-frozen epithelial OCs and evaluated a putative biological role of both representatives of the two antagonistic chromatin remodeling complexes by analyzing their associations with clinico-pathological characteristics and using data on patients’ survival as benchmarks.
Results
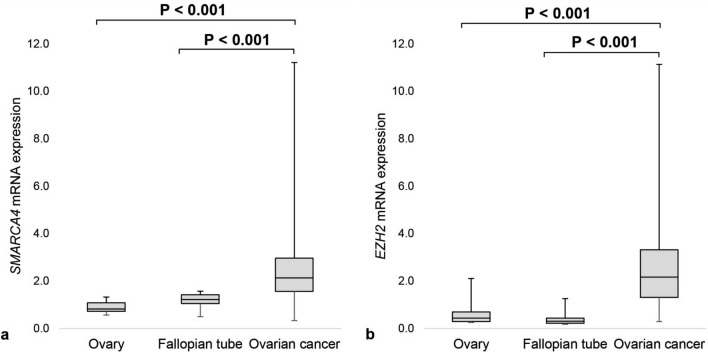
We analyzed mRNA expression of SMARCA4 and EZH2 in 238 OC, nineteen samples of non-neoplastic and otherwise histologically normal fallopian tube mucosa and sixteen histologically normal ovarian tissues. Median SMARCA4 and EZH2 mRNA levels in OC tissues compared to normal fallopian tubes were 1.7-fold and 7.2-fold higher, respectively (P < 0.001 for both) and 2.6-fold and 5.1-fold higher compared to non-neoplastic ovarian tissues (P < 0.001 for both) (Fig. 1).
Figure 1.
SMARCA4 and EZH2 mRNA expression in ovarian tissues. (a) SMARCA4 mRNA expression in 16 non-neoplastic ovaries, 19 non-neoplastic fallopian tubes and 238 OC tissues, (b) EZH2 mRNA expression in 16 non-neoplastic ovaries, 19 non-neoplastic fallopian tubes and 238 OC tissues.
While no relevant differences in SMARCA4 expression between the various histological OC subtypes were found, significant differences between histological subtypes were pointed out in EZH2 mRNA expression (P < 0.001). In detail, highest levels of EZH2 expression were found in the pooled group of high-grade serous (HGSOC) and endometrioid (HGEOC) cancers (P < 0.001). However, it is noteworthy, that in endometrioid cancers EZH2 mRNA levels were essentially the same regardless whether cancers were assigned to high-grade or low-grade cancers. This was not the case in cancers with serous histology, where significantly lower EZH2 levels were found in low-grade serous (LGSOC), as compared with high-grade serous OCs (P < 0.001); (Table 1).
Table 1.
Association of SMARCA4 and EZH2 mRNA expression with clinico-pathological characteristics in 238 OC patients.
| Variable | Number (%) | mRNA expression values (arbitrary units) | |||
|---|---|---|---|---|---|
| SMARCA4 | EZH2 | ||||
| Median (IQR) | P value | Median (IQR) | P value | ||
| Age | |||||
| ≤ 62 years (median age) | 120 (50.4%) | 2.250 (1.74–3.35) | 0.064 | 2.135 (1.24–3.90) | 0.487 |
| > 62 years | 118 (49.6%) | 2.056 (1.46–2.85) | 2.185 (1.32–3.03) | ||
| ≤ 51.5 years | 53 (22.3%) | 2.187 (1.63–3.54) | 0.381 | 1.660 (0.75–3.91) | 0.060 |
| > 51.5 years | 185 (77.7%) | 2.099 (1.56–2.87) | 2.300 (1.49–3.24) | ||
| ≤ 70 years | 167 (70.2%) | 2.219 (1.71–3.20) | 0.028 | 2.270 (1.41–3.73) | 0.168 |
| > 70 years | 71 (29.8%) | 2.005 (1.32–2.81) | 1.940 (1.25–2.94) | ||
| ≤ 40 years | 14 (5.9%) | 1.961 (1.13–2.46) | 0.151 | 1.155 (0.66–1.90) | 0.004 |
| > 40 years | 224 (94.1%) | 2.163 (1.61–3.04) | 2.255 (1.47–3.36) | ||
| Histology | |||||
| HGSOC | 144 (60.5%) | 2.250 (1.72–3.07) | 0.695 | 2.390 (1.51–3.69) | < 0.001 |
| LGSOC | 12 (5.0%) | 1.876 (1.53–2.48) | 0.710 (0.56–1.52) | ||
| HGEOC | 31 (13.0%) | 2.392 (1.15–3.98) | 2.530 (1.78–5.39) | ||
| LGEOC | 13 (5.5%) | 2.093 (1.76–2.55) | 2.520 (1.59–3.16) | ||
| Clear cell OC | 11 (4.6%) | 2.216 (1.13–3.33) | 1.310 (0.86–2.88) | ||
| Mucinous OC | 27 (11.3%) | 2.123 (1.44–3.08) | 1.570 (0.80–2.56) | ||
| FIGO stage | |||||
| I/II | 64 (26.9%) | 2.313 (1.58–3.35) | 0.193 | 2.125 (1.27–3.10) | 0.659 |
| III/IV | 174 (73.1%) | 2.082 (1.57–2.89) | 2.185 (1.39–3.41) | ||
| Tumor grade | |||||
| Low grade | 30 (12.6%) | 1.915 (1.55–2.47) | 0.145 | 1.410 (0.68–2.54) | 0.001 |
| Grade III | 208 (87.4%) | 2.219 (1.58–3.09) | 2.320 (1.48–3.58) | ||
| Residual disease | |||||
| Complete debulking | 114 (47.9%) | 2.219 (1.69–3.37) | 0.044 | 2.125 (1.19–3.29) | 0.647 |
| Any residual | 114 (47.9%) | 2.076 (1.48–2.76) | 2.185 (1.48–3.33) | ||
| Not indicated | 10 (4.2%) | – | – | ||
| Subgroups | |||||
| “Low-grade OC” | 25 (11.8%) | 1.917 (1.61–2.50) | 0.188 | 1.590 (0.71–2.61) | 0.006 |
| “High-grade OC” | 186 (88.2%) | 2.250 (1.65–3.08) | 2.410 (1.52–3.87) | ||
| BRCA1 mutation status | |||||
| Wild-type | 155 (65.1%) | 2.093 (1.64–2.87) | 0.587 | 2.060 (1.25–3.22) | 0.013 |
| BRCA1 mutated | 35 (14.7%) | 2.437 (1.79–3.20) | 3.070 (1.90–4.51) | ||
| Not indicated | 48 (20.2%) | – | – | ||
| BRCA2 mutation status | |||||
| Wild-type | 181 (76.0%) | 2.111 (1.65–2.89) | 0.787 | 2.240 (1.41–3.29) | 0.313 |
| BRCA2 mutated | 9 (3.8%) | 1.904 (1.55–3.37) | 3.910 (1.10–7.05) | ||
| Not indicated | 48 (20.2%) | – | – | ||
IQR interquartile range.
Bold values indicate P < 0.05.
SMARCA4 and EZH2 transcript levels related to clinico-pathological characteristics of OC patients
Associations of SMARCA4 and EZH2 expression with various clinico-pathological parameters are summarized in Table 1. Subgroup analyses are shown in Supplementary Table S1 and S2.
No association with age at time of diagnosis was revealed for either SMARCA4 or EZH2 expression when median age in the collective (62 years) or the median age at menopause in Europe (51.5 years)30 was used as a cut-off. Nonetheless, we have been interested whether the expression of both antagonistic variables is equally distributed independently of age or linked to specific age groups. We revealed significantly lower SMARCA4 mRNA expression in patients older than 70 years at the time of diagnosis (P = 0.028) and in contrast, lower EZH2 expression in young patients under 40 years of age (P = 0.004).
In “high-grade OCs” elevated SMARCA4 transcripts were associated with complete resection after surgical debulking (P = 0.016) (Supplementary Table S1).
In contrast, EZH2 expression was higher in the subgroup of “high-grade OCs” (P = 0.006). Furthermore, there was a significantly higher EZH2 mRNA expression in cancers with confirmed BRCA1 mutations (P = 0.013). However, no significant difference in EZH2 transcript levels was related to the BRCA2 mutational status, but it is worth to note that the number of included cases with BRCA2 mutations was very low (n = 9).
In the whole OC cohort, a strong positive correlation between SMARCA4 and EZH2 expression was uncovered (rs = 0.392, P < 0.001). Furthermore, both were directly associated with BRCA1 and BRCA2 mRNA expression and with cell cycle promoting regulator E2F3a, whereas EZH2 was additionally associated with the cell cycle promotor E2F1 (Supplementary Table S3).
Survival analysis related to the expression of SMARCA4 and EZH2
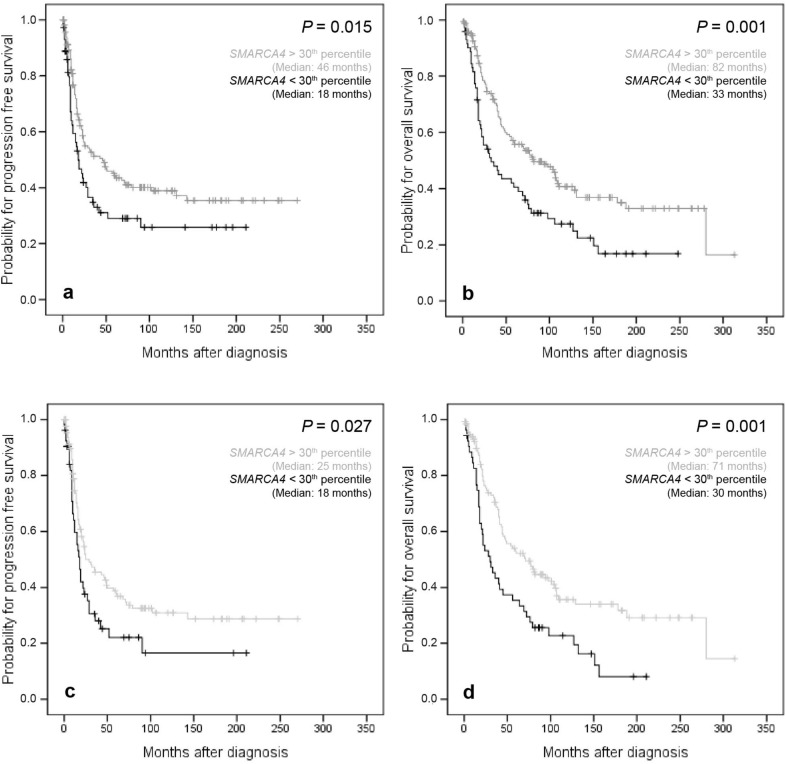
Univariate survival analysis in the entire cohort of OC patients showed that high SMARCA4 mRNA expression was associated with a favorable PFS (P = 0.015) and OS (P = 0.001). The same was especially true for the subgroup of “high-grade OCs” (P = 0.027 and P = 0.001, respectively) (Fig. 2). In “low-grade OCs” no clinical prognostic impact could be found for either PFS or OS which could be caused by the low number of included samples (n = 30).
Figure 2.
Kaplan–Meier survival analysis and SMARCA4 mRNA expression according to the 30th percentile as cut off-value in the whole cohort and in the subgroup of “high-grade OC” patients. (a) Progression free survival in 238 OC patients, (b) overall survival in 238 OC patients, (c) progression free survival in 186 “high-grade OC” patients, (d) overall survival in 186 “high-grade OC” patients.
In multivariate Cox-regression analysis SMARCA4 mRNA expression retained independent prognostic significance for both PFS and OS when the whole collective of OCs was considered (HR 0.66, P = 0.03 and HR 0.65, P = 0.018, respectively) (Table 2). However, in the subgroup of “high-grade OCs” independency of prognostic impact was only disclosed for OS (HR 0.64, P = 0.025) (Table 3).
Table 2.
Multivariate survival analysis in 238 OC patients. Analysis in (A) EZH2 high expressing cancers and (B) EZH2 ultra-high expressing cancers.
| Variable | Progression free survival | Overall survival | |||
|---|---|---|---|---|---|
| HR (95% CI) | P value | HR (95% CI) | P value | ||
| A: EZH2 high expressing cancers | |||||
| Age | Low versus high | 1.63 (1.11–2.39) | 0.013 | 2.49 (1.74–3.56) | < 0.001 |
| FIGO stage | I/II versus III/IV | 2.74 (1.49–5.03) | 0.001 | 1.44 (0.84–2.47) | 0.188 |
| Grading | Low grade versus grade III | 1.74 (0.76–3.95) | 0.189 | 2.59 (1.09–6.13) | 0.031 |
| Residual disease | No versus yes | 2.2 (1.44–3.35) | < 0.001 | 2.5 (1.63–3.84) | < 0.001 |
| SMARCA4 (30th percentile) | Low versus high | 0.66 (0.45–0.96) | 0.03 | 0.65 (0.46–0.93) | 0.018 |
| EZH2 (29th percentile) | Low versus high | 1.62 (1.04–2.52) | 0.034 | 1.23 (0.83–1.82) | 0.305 |
| B: EZH2 ultra-high expressing cancers | |||||
| Age | Low versus high | 1.54 (1.05–2.25) | 0.027 | 2.42 (1.70–3.45) | < 0.001 |
| FIGO stage | I/II versus III/IV | 2.98 (1.64–5.43) | < 0.001 | 1.57 (0.91–2.69) | 0.102 |
| Grading | Low grade versus grade III | 2.32 (1.05–5.10) | 0.037 | 2.92 (1.25–6.81) | 0.013 |
| Residual disease | No versus yes | 2.14 (1.40–3.27) | < 0.001 | 2.38 (1.55–3.65) | < 0.001 |
| SMARCA4 (30th percentile) | Low versus high | 0.81 (0.56–1.17) | 0.26 | 0.73 (0.51–1.04) | 0.079 |
| EZH2 (94th percentile) | Low versus high | 0.21 0.07–0.68) | 0.009 | 0.4 (0.16–1.01) | 0.032 |
HR hazard ratio, CI confidence interval.
Bold values indicate P < 0.05. The significance level was determined by Cox regression analysis.
Table 3.
Multivariate survival analysis in 186 “high-grade OC” patients. Analysis in (A) EZH2 high expressing cancers and (B) EZH2 ultra-high expressing cancers.
| Variable | Progression free survival | Overall survival | |||
|---|---|---|---|---|---|
| HR (95% CI) | P value | HR (95% CI) | P value | ||
| A: EZH2 high expressing cancers | |||||
| Age | Low versus high | 1.74 (1.14–2.66) | 0.010 | 2.40 (1.61–3.58) | < 0.001 |
| FIGO stage | I/II versus III/IV | 2.06 (1.03–4.14) | 0.041 | 1.15 (0.62–2.13) | 0.658 |
| Residual disease | No versus yes | 2.51 (1.56–4.06) | < 0.001 | 2.68 (1.64–4.37) | < 0.001 |
| SMARCA4 (30th percentile) | Low versus high | 0.76 (0.50–1.15) | 0.189 | 0.64 (0.43–0.95) | 0.025 |
| EZH2 (29th percentile) | Low versus high | 1.67 (1.01–2.74) | 0.045 | 1.45 (0.92–2.28) | 0.108 |
| B: EZH2 ultra-high expressing cancers | |||||
| Age | Low versus high | 1.66 (1.09–2.52) | 0.017 | 2.28 (1.54–3.37) | < 0.001 |
| FIGO stage | I/II versus III/IV | 2.50 (1.25–5.00) | 0.010 | 1.32 (0.71–2.45) | 0.378 |
| Residual disease | No versus yes | 2.36 (1.46–3.80) | < 0.001 | 2.46 (1.51–4.01) | < 0.001 |
| SMARCA4 (30th percentile) | Low versus high | 0.95 (0.63–1.44) | 0.819 | 0.74 (0.50–1.09) | 0.124 |
| EZH2 (94th percentile) | Low versus high | 0.20 (0.06–0.65) | 0.007 | 0.43 (0.17–1.07) | 0.069 |
HR hazard ratio.
Bold values indicate P < 0.05. The significance level was determined by Cox regression analysis.
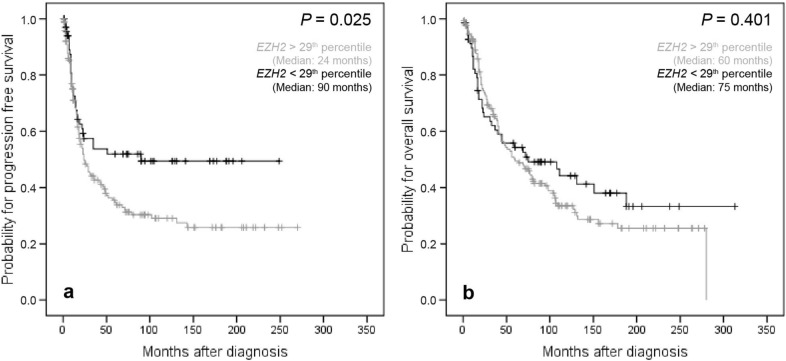
As Youden’s threshold determination for EZH2 yielded a S-shaped ROC-curve, two cut-off points (29th and 94th percentile) predicting opposite features were defined. In contrast to SMARCA4 high EZH2 mRNA levels proved to be associated with impaired PFS (P = 0.025) in the entire OC collective when the threshold for dichotomization between high and low was set at the 29th percentile. However, for OS no clinical prognostic impact was delineated in univariate survival analysis at this threshold (Fig. 3). Moreover, in multivariate analysis the prognostic impact of EZH2 transcript levels could be confirmed for PFS in the whole collective of OCs (HR 1.62, P = 0.034) (Table 2) and in the subgroup of “high-grade OCs” (HR 1.67, P = 0.045) (Table 3). No independent impact of EZH2 mRNA levels on clinical outcome was revealed when the subgroup of “low-grade OCs” was analyzed separately for PFS and OS.
Figure 3.
Kaplan–Meier survival analysis and EZH2 mRNA expression according to the 29th percentile as cut-off value in the whole cohort. (a) Progression free survival in 238 OC patients, (b) overall survival in 238 OC patients.
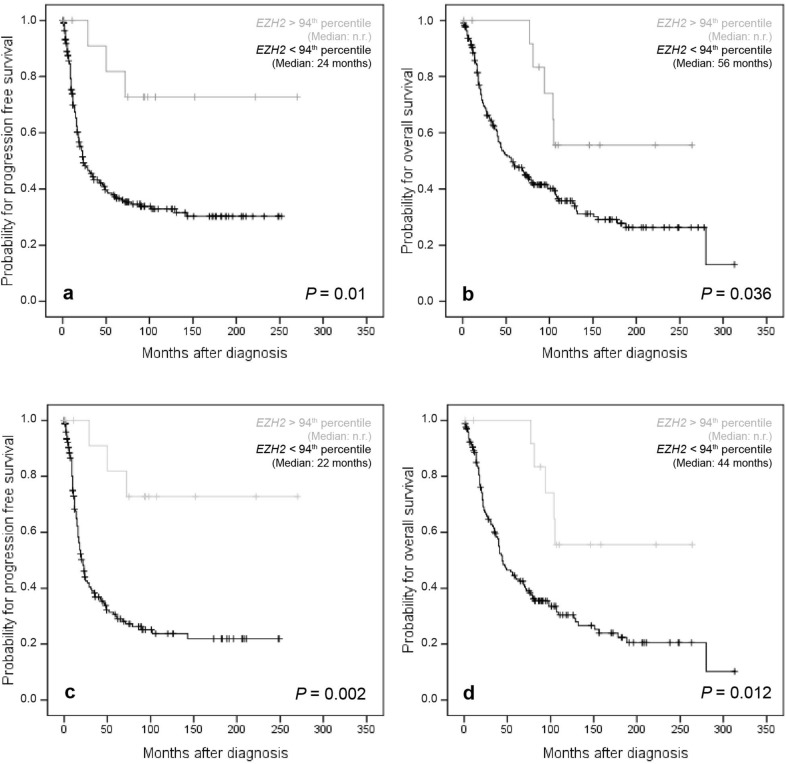
However, when the 94th percentile was used for cohort dichotomization, a conversion of the prognostic attribute of EZH2 expression from unfavorable to favorable clinical outcome became apparent for both the PFS and OS. Thus, with these percentiles very high EZH2 mRNA levels were associated with improved PFS (P = 0.01) and OS (P = 0.036) in the whole investigated cohort and in the subgroup of “high-grade OCs” (P = 0.002 and P = 0.012, respectively) (Fig. 4). It is worth noting that all tumors classified as ultra-high EZH2 expressing were all high-grade cancers. Moreover, Cox-regression analysis confirmed the independency of prognostic relevance of ultra-high EZH2 mRNA expression for PFS and OS in the whole cohort (HR 0.21, P = 0.009 and HR 0.4, P = 0.032, respectively) (Table 2) and for PFS in the subgroup of “high-grade OCs” (HR 0.2, P = 0.007) (Table 3).
Figure 4.
Kaplan–Meier survival analysis and EZH2 mRNA expression according to the 94th percentile as cut off-value in the whole cohort and in the subgroup of “high-grade OC” patients. (a) Progression free survival in 238 OC patients, (b) overall survival in 238 OC patients, (c) progression free survival in 186 “high-grade OC” patients, (d) overall survival in 186 “high-grade OC” patients; n.r. not reached.
Validation of SMARCA4 and EZH2 survival analysis in independent cohorts
Survival analyses employing mRNA expression data from the Niigata OC cohort (n = 110) and the MSKCC OC cohort (n = 185) confirmed the prognostic relevance of SMARCA4. High SMARCA4 mRNA expression was associated with a better survival (Niigata cohort: PFS: P = 0.011, OS: P = 0.014; MSKCC cohort: PFS: P = 0.003; OS: P = 0.009; Supplementary Fig. S1).
Prognostic relevance was also confirmed for EZH2, where high EZH2 mRNA expression was associated with poor PFS in the Niigata cohort (cut-off: 12th percentile; P = 0.048), which in contrast to our cohort was translated in impaired OS (cut-off: 12th percentile; P = 0.015). The latter finding was corroborated by the Tothill cohort (cut-off: 25th percentile P = 0.020) (Supplementary Fig. S2).
Extremely high cut-offs used for cohort dichotomization revealed no conversion of the prognostic attribute of EZH2 expression as observed in our data. Interestingly, however, in the Duke cohort high EZH2 mRNA expression was associated with an improved OS (P = 0.008) when a high EZH2 expression cut-off (69th percentile) was used (Supplementary Fig. S2).
Discussion
In the present work expression of SMARCA4 and EZH2 as two key players of the functionally antagonist DNA remodeling complexes SWI/SNF and PRC2, respectively, were analyzed on the transcriptome level in OCs. Whereas the SWI/SNF complex opens accessibility to chromatin, especially to tumor-suppressor genes, by removal of PCR2 and promotes cellular differentiation, the PRC2 complex, in contrast, stalls differentiation and favors malignant transformation. Imbalances between both complexes can be due to overexpression of EZH2 in undifferentiated cancers, but most commonly occur in cancers by mutations of members of the SWI/SNF complex, such as ARID1A and SMARCA4 that together with SMARCA2 is the ATPase subunit of this tumor-suppressing complex. Such imbalances lead to a supremacy of EZH2, the catalytic subunit of PRC2, which is the exclusive human methyltransferase for H3K27. Aberrant tri-methylated H3K27 represents a transcriptionally repressive histone mark that is highly oncogenic in a broad spectrum of human cancers.
This dualistic behavior regarding both complexes is well reflected in the herein performed expression analyses of SMARCA4 and EZH2 in terms that high expression of SMARCA4 independently reflects a favorable PFS and OS whereas elevated EZH2 was independently associated with improved clinical outcome. Moreover, high SMARCA4 mRNA levels are associated with better surgical resectability, especially in high-grade cancers. This is of importance, since the residual disease after primary debulking surgery is the most important prognostic factor in OC and is often wrongly attributed to surgical skills only. Albeit associated with cell-cycle promoting E2F3a, a known prognosticator in OC31, high SMARCA4 expressing cancers were associated with improved survival. However, Guerrero-Martinez and Reyes showed high expression of SMARCA4 to be associated with worse prognosis in OC32. These findings are not only in conflict with our data but also with those of the Niigata OC cohort and the MSKCC cohort.
Our data show that high EZH2 transcript levels were related to high-grade, BRCA1-mutated ovarian cancers, which in turn were very strongly associated with cell cycle promoting factors E2F1 and E2F3a. All this indicates that EZH2 high expressing cancers exhibit a very high proliferative turnover. A fact that is additionally underscored by the significant relationship with the expression of BRCA1 and -2 mRNA, which is thought to reflect high DNA repair events known to occur at higher rates in tumors with high mitotic activity33. Thus, it is not unexpected that overexpression of growth promoting and oncogenic EZH2 is associated with impaired PFS in univariate and multivariate survival analysis. Accordingly, elevated EZH2 expression was also associated with adverse prognosis in several other tumor entities, such as breast, cervical, larynx and colorectal cancers34–37. Rao et al. also showed that high EZH2 expression was associated with poor prognosis in OC and observed a positive correlation between overexpression of EZH2 and TGF-β1 suggesting a potential role of EZH2 in the control of cell migration and invasion via regulation of TGF-β1 expression38. A further hint for EZH2 promoting effects on migration and invasion was provided by Yi et al. who showed that EZH2 inhibits the expression of the physiological matrix metalloproteinase inhibitor TIMP2 in ovarian cancer cells39. Moreover, in OC EZH2 upregulation via inhibition of its negative regulator miRNA-137 by c-Myc has been implicated into promotion of cisplatin resistance40.
However, due to the S-shaped ROC curve in the calculation of the Youden’s Index we herein uncovered that at very high EZH2 transcript levels the effect on survival converted from unfavorable to favorable PFS and OS. In contrast to our findings of poor prognosis at lower threshold expression, outcome improvements were not only observed for PFS but also for OS and were furthermore statistically confirmed in the multivariate Cox-regression analysis. We hypothesize that this conversion is not due simply to statistical unevenness but may be the result of different biological effects, which occur after a critical EZH2 activity is reached in the concerned tumor cells. Very recently, the inhibitory role of EZH2 on the activity of MAD2L2 (Rev7), an essential subunit of the shieldin complex, was highlighted. Shieldin/Rev7 protects DNA broken ends from a 5′-3′ resection at double strand breaks (DSB), which is essential that RAD51-dependent homologous recombination (HR) repair can occur29. Thus, ultra-high EZH2 activity favors HR repair over non-homologous end-joining (NHEJ), by lowering Rev7 activity. In line with this, Xu et al.41 reported that mutation of Rev7 or abrogation of its activity was found to increase HR capacity and to confer resistance to PARP inhibitors on the one hand but hypersensitivity toward platinum containing drugs on the other hand41–43. As almost all of the herein included patients were treated with platinum-based chemotherapy but none of them were treated with a PARP inhibitor in any of the lines of treatment, it is conceivable that the observed survival advantage in the small percentage of patients with ultra-high EZH2 expressing tumors may originate from improved responsiveness to platinum-based treatment. Detailed analysis of the EZH2 ultra-high cohort revealed that all these cancers in fact, exhibited platinum-hypersensitivity and were without exception advanced high-grade tumors. For comparison, in the stage-adjusted cohort of high-grade cancers with lower EZH2 expression, the rate of platinum-hypersensitivity was as low as 17.5% (data not shown). In accordance, loss of EZH2 has been recently described to drive resistance to carboplatin and paclitaxel in serous OCs provided ATM is upregulated44. These findings should give cause for concern regarding an indiscriminative use of EZH2 inhibitors combined with platinum-based chemotherapy in OC patients.
EZH2 is a good example showing that a biomarker can change its prognostic attributes with the levels of its expression in the same tumor entity. Such conversions may be dependent on a specific molecular context in the concerned cancers, for instance regarding EZH2 this could be either a concomitant overexpression of ATM or changing features in the interplay with Rev729,44.
Methods
Patients and samples
Ovarian tissue samples from 238 patients with epithelial OCs were obtained at primary debulking surgery in the period from 2006 to 2017. Patients were 22–90 years old and the median age at the time of diagnosis was 62 years. Additionally, nineteen non-neoplastic fallopian tubal tissue specimens and 16 non-neoplastic ovarian tissues were used as a control group. These samples were obtained from elective salpingo-oophorectomies for benign conditions (i.e. salpingectomy for sterilization or contralateral salpingo-oophorectomy as part of surgery for benign cysts). Median age at time of surgery was 52 years (range 19–81 years). The tissue probes were collected and processed at the Pathology Unit of the Department of Obstetrics and Gynecology of the Medical University of Innsbruck, Austria.
Written informed consent was obtained from all patients before enrolment. The study was reviewed and approved by the Ethics committee of the Medical University of Innsbruck (reference number: 1157/2018) and conducted in accordance with the Declaration of Helsinki. All samples were anonymized before the analysis. The median follow-up period was 20 months (IQR 9–73 months) regarding the progression-free survival and 45 months (IQR 18–107 months) concerning overall survival. Clinico-pathological characteristics of included patients are listed in Table 1.
Analyses were performed in the whole OC cohort as well as in two subgroups— “high-grade OCs” and “low-grade OCs”, separately. For the latter evaluations, the final subdivision was made in accordance with the WHO classification of 2020 by a second pathologic review45. In these subgroup analyses mucinous cancers were excluded.
Quantitative real time PCR
Total cellular RNA extraction from and reverse transcription were performed as previously described33.
Primers and probes for the TATA box-binding protein (TBP; a component of the DNA-binding protein complex TFIID as an endogenous RNA control) were used according to Bieche et al.46. Primers and probes for EZH2 and SMARCA4 were purchased from Applied Biosystems (Foster City, CA, USA, Applied Biosystems Assay ID: Hs00544830_m1 and Hs00231324_m1). PCR reactions were performed as previously described33. The standard curves were generated using serially diluted solutions of standard cDNA derived from the HTB-77 carcinoma cell line. Each experiment included this standard curve, a positive control sample (OVCAR3 carcinoma cell-line for EZH2; HOC7 for SMARCA4), 40 patient samples and a no template control. Real-time PCR assays were conducted in duplicates for each sample, and the mean value was used for calculation. The expression analyses of BRCA1, BRCA2, E2F1 and E2F3a were recently described31,33.
Data normalization was carried out against TBP expression, the endogenous RNA-control and expressed in arbitrary units.
Mutation analysis
Genomic DNA from pulverized, quick-frozen OC specimens was isolated using the DNeasy tissue-kit (Qiagen, Hilden, Germany). Targeted NGS was performed using the TruSight Cancer sequencing panel (Illumina, San Diego, USA) as described recently33.
Validation cohort
To validate findings from our study cohort, we employed gene expression data from independent cohorts of OC patients publically available at GEO, a public functional genomics data repository (GSE17260 (Niigata, Yoshihara cohort), GSE9891 (AOCS, RBH, WH, NKI-AVL, Tothill cohort), GSE26712 (MSKCC, Bonome cohort)) and available via PrognoScan, a new database for meta-analysis of the prognostic value of genes (DUKE-OC, Bild cohort)47.
Statistical analysis
The non-parametric Mann–Whitney U test or Kruskal–Wallis test were applied to test for statistical significance between two groups or more than two groups, respectively. To assess the correlations between SMARCA4 mRNA-expression, EZH2 mRNA-expression and different other molecular markers Spearman-rank correlation analyses were used.
Progression-free survival (PFS) was defined as the time from diagnosis of the primary tumor to the histopathological confirmation of recurrence. Overall survival (OS) was defined as the time from diagnosis to death from any cause or to the last clinical inspection. Univariate Kaplan–Meier analyses and multivariate Cox-regression survival analyses were used to explore the association of SMARCA4 mRNA expression and EZH2 mRNA expression with PFS and OS.
In order to investigate the biological impact of SMARCA4 and EZH2 expression on the basis of the clinical outcome in the cohort, Youden’s Index48 was used to identify the optimal threshold for both parameters to distinguish between “low” and “high” expression. The optimal discriminatory cut-off point for SMARCA4 corresponded to the 30th percentile. As for EZH2 the Receiver Operating Characteristics (ROC) curve was S-shaped and crossed the diagonal random classifier line, which separates both areas of opposite characteristics, two discriminatory cut-off points were defined for EZH2: the one at the 29th and the second at the 94th percentile.
P values < 0.05 were considered as statistically significant. Statistical analysis was performed using SPSS statistical software (version 20.0.0; SPSS Inc., Chicago, IL, USA).
Ethics approval
The study was reviewed and approved by the Ethics committee of the Medical University of Innsbruck (reference number: 1157/2018) and conducted in accordance with the Declaration of Helsinki.
Consent for publication
Written informed consent was obtained from all patients before enrolment.
Supplementary information
Acknowledgements
We thank Annemarie Wiedemair, Julia Rössler and Inge Gaugg for their excellent technical assistance.
Author contributions
A.G.Z., H.F. and C.M. developed the study concept, A.G.Z., H.F., C.M. and K.L. designed the project and edited the manuscript, K.L., I.T., V.W., K.K., and D.R. were involved in data acquisition, K.L., I.T. and H.F. performed statistical analyses, K.L. and A.G.Z. prepared the manuscript, A.G.Z., H.F., C.M., K.L., I.T., V.W., K.K., and D.R. analyzed and interpreted data and reviewed the final manuscript.
Funding
The project was supported by the Verein zur Krebsforschung in der Frauenheilkunde, an association which is exclusively financed by donation funds for cancer research in female malignancies (Grant Number 2365/18).
Data availability
The source dataset is shown in Supplementary Table S4.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-77532-x.
References
- 1.Webb PM, Jordan SJ. Epidemiology of epithelial ovarian cancer. Best Pract. Res. Clin. Obstet. Gynaecol. 2017;41:3–14. doi: 10.1016/j.bpobgyn.2016.08.006. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J. Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 3.Cannistra SA. Cancer of the ovary. N. Engl. J. Med. 2004;351:2519–2529. doi: 10.1056/NEJMra041842. [DOI] [PubMed] [Google Scholar]
- 4.Kadoch C, Copeland RA, Keilhack H. PRC2 and SWI/SNF chromatin remodeling complexes in health and disease. Biochemistry. 2016;55:1600–1614. doi: 10.1021/acs.biochem.5b01191. [DOI] [PubMed] [Google Scholar]
- 5.Poynter ST, Kadoch C. Polycomb and trithorax opposition in development and disease. Wiley Interdiscip. Rev. Dev. Biol. 2016;5:659–688. doi: 10.1002/wdev.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilson BG, Roberts CW. SWI/SNF nucleosome remodellers and cancer. Nat. Rev. Cancer. 2011;11:481–492. doi: 10.1038/nrc3068. [DOI] [PubMed] [Google Scholar]
- 7.Shain AH, Pollack JR. The spectrum of SWI/SNF mutations, ubiquitous in human cancers. PLoS ONE. 2013;8:e55119. doi: 10.1371/journal.pone.0055119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kadoch C, et al. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat. Genet. 2013;45:592–601. doi: 10.1038/ng.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wiegand KC, et al. ARID1A mutations in endometriosis-associated ovarian carcinomas. N. Engl. J. Med. 2010;363:1532–1543. doi: 10.1056/NEJMoa1008433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wiegand KC, et al. Loss of BAF250a (ARID1A) is frequent in high-grade endometrial carcinomas. J. Pathol. 2011;224:328–333. doi: 10.1002/path.2911. [DOI] [PubMed] [Google Scholar]
- 11.Shao F, et al. Clinicopathological significance of ARID1B in breast invasive ductal carcinoma. Histopathology. 2015;67:709–718. doi: 10.1111/his.12701. [DOI] [PubMed] [Google Scholar]
- 12.Varela I, et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature. 2011;469:539–542. doi: 10.1038/nature09639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Versteege I, et al. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature. 1998;394:203–206. doi: 10.1038/28212. [DOI] [PubMed] [Google Scholar]
- 14.Reisman DN, Sciarrotta J, Wang W, Funkhouser WK, Weissman BE. Loss of BRG1/BRM in human lung cancer cell lines and primary lung cancers: correlation with poor prognosis. Cancer Res. 2003;63:560–566. [PubMed] [Google Scholar]
- 15.Scully, R. E. Tumors of the Ovary and Maldeveloped Gonads (Armed Forces Institute of Pathology, 1979).
- 16.Witkowski L, et al. Germline and somatic SMARCA4 mutations characterize small cell carcinoma of the ovary, hypercalcemic type. Nat. Genet. 2014;46:438–443. doi: 10.1038/ng.2931. [DOI] [PubMed] [Google Scholar]
- 17.Jelinic P, et al. Recurrent SMARCA4 mutations in small cell carcinoma of the ovary. Nat. Genet. 2014;46:424–426. doi: 10.1038/ng.2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y, et al. The histone methyltransferase EZH2 is a therapeutic target in small cell carcinoma of the ovary, hypercalcaemic type. J. Pathol. 2017;242:371–383. doi: 10.1002/path.4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martinez-Borges AR, et al. Familial small cell carcinoma of the ovary. Pediatr. Blood Cancer. 2009;53:1334–1336. doi: 10.1002/pbc.22184. [DOI] [PubMed] [Google Scholar]
- 20.Young RH, Oliva E, Scully RE. Small cell carcinoma of the ovary, hypercalcemic type. A clinicopathological analysis of 150 cases. Am. J. Surg. Pathol. 1994;18:1102–1116. doi: 10.1097/00000478-199411000-00004. [DOI] [PubMed] [Google Scholar]
- 21.Bracken AP, Helin K. Polycomb group proteins: navigators of lineage pathways led astray in cancer. Nat. Rev. Cancer. 2009;9:773–784. doi: 10.1038/nrc2736. [DOI] [PubMed] [Google Scholar]
- 22.Kleer CG, et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc. Natl. Acad. Sci. USA. 2003;100:11606–11611. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bachmann IM, et al. EZH2 expression is associated with high proliferation rate and aggressive tumor subgroups in cutaneous melanoma and cancers of the endometrium, prostate, and breast. J. Clin. Oncol. 2006;24:268–273. doi: 10.1200/JCO.2005.01.5180. [DOI] [PubMed] [Google Scholar]
- 24.Varambally S, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 25.Lee SR, et al. Activation of EZH2 and SUZ12 regulated by E2F1 predicts the disease progression and aggressive characteristics of bladder cancer. Clin. Cancer Res. 2015;21:5391–5403. doi: 10.1158/1078-0432.CCR-14-2680. [DOI] [PubMed] [Google Scholar]
- 26.Li H, Cai Q, Godwin AK, Zhang R. Enhancer of zeste homolog 2 promotes the proliferation and invasion of epithelial ovarian cancer cells. Mol. Cancer Res. 2010;8:1610–1618. doi: 10.1158/1541-7786.MCR-10-0398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Italiano A, et al. Tazemetostat, an EZH2 inhibitor, in relapsed or refractory B-cell non-Hodgkin lymphoma and advanced solid tumours: a first-in-human, open-label, phase 1 study. Lancet Oncol. 2018;19:649–659. doi: 10.1016/S1470-2045(18)30145-1. [DOI] [PubMed] [Google Scholar]
- 28.Kurmasheva RT, et al. Initial testing (stage 1) of tazemetostat (EPZ-6438), a novel EZH2 inhibitor, by the Pediatric Preclinical Testing Program. Pediatr. Blood Cancer. 2017;64:e26218. doi: 10.1002/pbc.26218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karakashev S, et al. EZH2 inhibition sensitizes CARM1-high, homologous recombination proficient ovarian cancers to PARP inhibition. Cancer Cell. 2020;37:157–167. doi: 10.1016/j.ccell.2019.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Palacios S, Henderson VW, Siseles N, Tan D, Villaseca P. Age of menopause and impact of climacteric symptoms by geographical region. Climacteric. 2010;13:419–428. doi: 10.3109/13697137.2010.507886. [DOI] [PubMed] [Google Scholar]
- 31.Reimer D, et al. E2F3a is critically involved in epidermal growth factor receptor-directed proliferation in ovarian cancer. Cancer Res. 2010;70:4613–4623. doi: 10.1158/0008-5472.CAN-09-3551. [DOI] [PubMed] [Google Scholar]
- 32.Guerrero-Martinez JA, Reyes JC. High expression of SMARCA4 or SMARCA2 is frequently associated with an opposite prognosis in cancer. Sci. Rep. 2018;8:2043. doi: 10.1038/s41598-018-20217-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsibulak I, et al. BRCA1 and BRCA2 mRNA-expression prove to be of clinical impact in ovarian cancer. Br. J. Cancer. 2018;119:683–692. doi: 10.1038/s41416-018-0217-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang X, et al. Clinical and prognostic relevance of EZH2 in breast cancer: a meta-analysis. Biomed. Pharmacother. 2015;75:218–225. doi: 10.1016/j.biopha.2015.07.038. [DOI] [PubMed] [Google Scholar]
- 35.Liu Y, et al. Increased EZH2 expression is associated with proliferation and progression of cervical cancer and indicates a poor prognosis. Int. J. Gynecol. Pathol. 2014;33:218–224. doi: 10.1097/PGP.0b013e31829c6574. [DOI] [PubMed] [Google Scholar]
- 36.Zhang MJ, et al. Clinical significance of USP7 and EZH2 in predicting prognosis of laryngeal squamous cell carcinoma and their possible functional mechanism. Int. J. Clin. Exp. Pathol. 2019;12:2184–2194. [PMC free article] [PubMed] [Google Scholar]
- 37.Abdel Raouf SM, Ibrahim TR, Abdelaziz LA, Farid MI, Mohamed SY. Prognostic value of TWIST1 and EZH2 expression in colon cancer. J. Gastrointest. Cancer. 2019 doi: 10.1007/s12029-019-00344-4. [DOI] [PubMed] [Google Scholar]
- 38.Rao ZY, et al. EZH2 supports ovarian carcinoma cell invasion and/or metastasis via regulation of TGF-beta1 and is a predictor of outcome in ovarian carcinoma patients. Carcinogenesis. 2010;31:1576–1583. doi: 10.1093/carcin/bgq150. [DOI] [PubMed] [Google Scholar]
- 39.Yi X, et al. EZH2-mediated epigenetic silencing of TIMP2 promotes ovarian cancer migration and invasion. Sci. Rep. 2017;7:3568. doi: 10.1038/s41598-017-03362-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun J, et al. miR-137 mediates the functional link between c-Myc and EZH2 that regulates cisplatin resistance in ovarian cancer. Oncogene. 2019;38:564–580. doi: 10.1038/s41388-018-0459-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu G, et al. REV7 counteracts DNA double-strand break resection and affects PARP inhibition. Nature. 2015;521:541–544. doi: 10.1038/nature14328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Konstantinopoulos PA, Ceccaldi R, Shapiro GI, D'Andrea AD. Homologous recombination deficiency: exploiting the fundamental vulnerability of ovarian cancer. Cancer Discov. 2015;5:1137–1154. doi: 10.1158/2159-8290.CD-15-0714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Niimi K, et al. Suppression of REV7 enhances cisplatin sensitivity in ovarian clear cell carcinoma cells. Cancer Sci. 2014;105:545–552. doi: 10.1111/cas.12390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Naskou J, et al. EZH2 loss drives resistance to carboplatin and paclitaxel in serous ovarian cancers expressing ATM. Mol. Cancer Res. 2020;18:278–286. doi: 10.1158/1541-7786.MCR-19-0141. [DOI] [PubMed] [Google Scholar]
- 45.McCluggage, W. G., Lax, S. F., Longacre, T. A., Malpica, A. & Soslow, R. A. WHO Classification of Tumors: Female Genital Tumours. 5 edn, Vol. 4 32–35 (2020).
- 46.Bieche I, Franc B, Vidaud D, Vidaud M, Lidereau R. Analyses of MYC, ERBB2, and CCND1 genes in benign and malignant thyroid follicular cell tumors by real-time polymerase chain reaction. Thyroid. 2001;11:147–152. doi: 10.1089/105072501300042802. [DOI] [PubMed] [Google Scholar]
- 47.Mizuno H, Kitada K, Nakai K, Sarai A. PrognoScan: a new database for meta-analysis of the prognostic value of genes. BMC Med. Genomics. 2009;2:18. doi: 10.1186/1755-8794-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Youden WJ. Index for rating diagnostic tests. Cancer. 1950;3:32–35. doi: 10.1002/1097-0142(1950)3:1<32::aid-cncr2820030106>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The source dataset is shown in Supplementary Table S4.