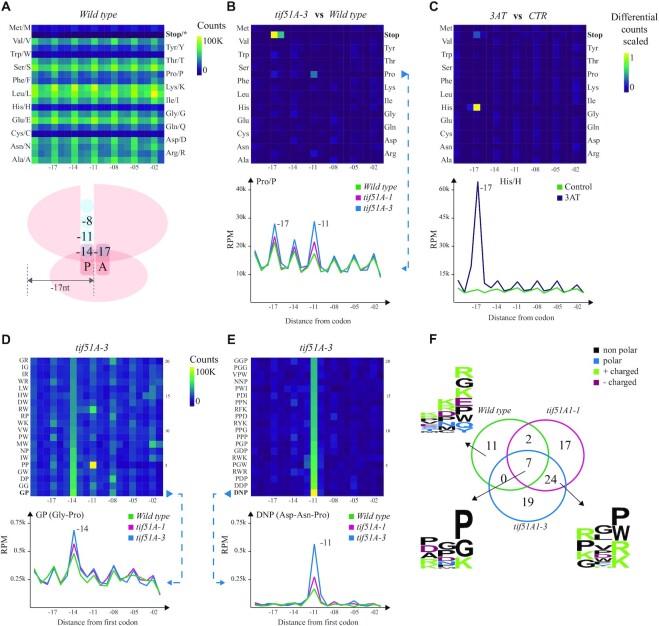
Figure 3.
Codon- and codon motif-specific ribosome protection patterns in budding yeast. (A) Heatmap of counts located at a certain distance from each amino acid in the wild type. The ribosome scheme depicts positions of 5′P endpoints relative to the ribosome. (B) A differential heatmap showing the difference in per row scaled 5′P counts between the tif5A1–3 eIF5A mutant and the wild type for all the amino acids, and a line chart highlighting the difference between both mutants and the wild type for proline (Pro/P). The x-axis shows the distance from the first base of a codon, and the y-axis represents RPM in the coding regions. (C) A differential heatmap of codon-specific 5′P counts between 3AT-treated and control yeast cells highlighting the stalling at −17 nt from histidine (His/H) and the corresponding line chart. (D) The top 20 dipeptides showing relatively high counts at −14 nt in the tif5A1–3 eIF5A mutant and the strongest pausing caused by GP (Gly–Pro) dipeptide, comparing the two eIF5A mutants and the wild type. (E) The top 20 tripeptide motifs showing the strongest −11 nt pausing in the tif5A1–3 eIF5A mutant and a line chart comparing the two mutants and the wild-type strains for the strongest pause-associated tripeptide DNP (Asp–Asn–Pro). (F) A Venn diagram showing the overlap of tripeptides with at least 3-fold increase in 5′P counts at position −11 nt compared to the background, in the three yeast strains. The logo plots present the motifs specific to the wild type, or the three strains or the two eIF5A mutants.