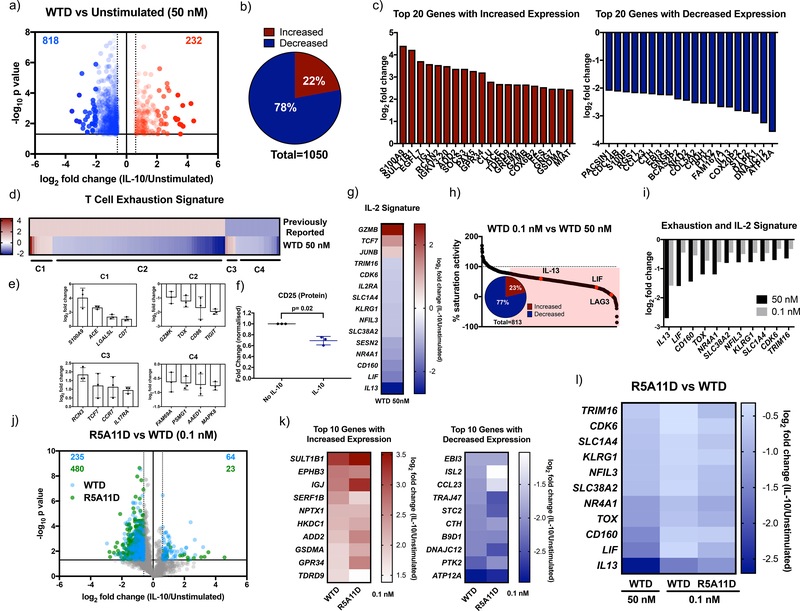
Figure 6. Characterisation of transcriptional activity induced by IL-10 and high affinity variants in human CD8+ T cells.
(a). Volcano plot of CD8+ T cell genes significantly increased by WTD 50 nM ≧ 0.6 log2 fold change (red) and significantly decreased ≦ −0.6 log2 fold change compared to unstimulated cells. Fold change was calculated by dividing 50 nM WTD values by unstimulated values for each donor. The average fold change was calculated and the log2 of this value is plotted. P values were calculated by two sample equal variance two-tailed t-test of the log2 fold change of 50 nM WTD/unstimulated genes for each donor. Genes that were not significantly changed (p>0.05) or were ≦ 0.6 ≧ −0.6 log2 fold change were excluded. (b). Proportion of CD8+ T cell genes whose expression was significantly increased or decreased by 50 nM WTD ≧ 0.6 or ≦ −0.6 log2 fold change compared to unstimulated cells. (c). Log2 fold change for the top 20 protein-coding genes whose expression was significantly increased (red) and decreased (blue) by 50 nM WTD in CD8+ T cells. (d). Heat map comparing a list of previously reported exhaustion-associated genes (43) to genes whose expression was significantly changed by 50 nM WTD. Previously reported genes were given a nominal value of 1 for genes with increased expression and −1 for genes with decreased expression. Log2 fold change for 50 nM WTD was plotted. Cluster 1 (C1) represents genes with increased expression in exhausted cells and with increased expression induced by 50 nM WTD. C2 represents genes with increased expression in exhausted cells and decreased expression induced by 50 nM WTD. C3 represents genes with decreased expression in exhausted cells and with increased expression induced by 50 nM WTD. C4 represents genes with decreased expression in exhausted cells and with increased expression induced by 50 nM WTD. (e). The log2 fold change induced by 50 nM WTD for selected genes from each cluster is shown. Each point represents one biological replicate. (f). Measurement of CD25 protein levels by flow cytometry in activated CD8+ T cells in a PBMC population in the presence or absence of IL-10. Fold change was calculated by dividing CD25 levels of IL-10 stimulated by non-IL-10 stimulated control values for each donor. Each point represents one biological replicate. P value was calculated by paired t test. (g). Heatmap showing the log2 fold change induced by 50 nM WTD for genes whose protein expression was previously reported to be altered by IL-2 in CD8+ T cells (44). (h). Percentage gene expression activity of low dose WTD (0.1 nM) compared to high dose WTD (50 nM). The log2 fold change of 0.1 nM WTD/unstimulated was divided by that for 50 nM WTD/unstimulated and multiplied by 100. Genes that showed ≦ 75% of high dose activity (813 genes) are highlighted in red. Insert shows the percentage of these genes with increased or decreased expression by WTD. (i). Log2 fold change for genes associated with exhaustion or IL-2 stimulation and their expression after treatment with WTD at 50 nM or 0.1 nM. (j). Volcano plot of genes whose expression was changed by WTD (blue) or R5A11D (green) at 0.1 nM in CD8+ T cells. Only genes that had already been shown to be significantly changed by 50 nM WTD are plotted. (k). Heatmap of the top 10 CD8+ T cell genes with increased or decreased expression by 0.1 nM WTD compared to 0.1 nM R5A11D. (l). Heatmap of exhaustion or IL-2 associated genes changed by WTD at 50 nM and 0.1 nM and R5A11D at 0.1 nM. CD8+ T cells from 3 donors were analysed in (a) to (l).