Abstract
Background:
Modern CT scanning can produce 4D images of the left atrial appendage (LAA). LAA function and morphology can then be measured, to plan interventions such as occlusion, and to evaluate LAA flow for thrombogenic risk analysis. A current problem here is defining a reproducible boundary between the LAA and the LA.
Methods:
This study used retrospectively gated 4D CT data from 25 implantation and coronary artery imaging patients. In each patient the LAA ostium was defined at multiple time points during the RR interval. In order to examine the reproducibility of the definition of the LAA ostium, 3 observers analyzed all time frames in each patient 3 times. Five non-consecutive time frames from each patient were then compared using intraclass correlation coefficients (ICC) to quantify the precision of the method across patients. The correlation of LAA volumes for each time frame of each patient was determined across the different observers (interobserver) and within each observer’s own datasets (intraobserver).
Results:
The method was successful in 92% of patients. Two-way random effect, absolute agreement, single measurement ICCs for interobserver measurements were 0.984, 0.990, and 0.988, with intraobserver ICCs of 0.989, 0.989, and 0.995. The ICC of all observations was 0.988.
Conclusions:
Classification of the LAA ostium using a stepwise procedure identifying the coumadin ridge and two vascular landmarks in ECG-gated CTA provides a viable method of establishing a highly reproducible boundary between the atrium and LAA needed to obtain LAA metrics useful for procedure planning and measuring LAA function.
Keywords: LAA, morphology, ostium, LA, CT, ICC
Subject Terms: Atrial Fibrillation, Physiology, Computerized Tomography (CT), Imaging, Catheter-Based Coronary and Valvular Interventions, Quality and Outcomes
Introduction
The left atrial appendage (LAA) is a highly trabeculated, actively contracting structure that has been implicated as the major source of thrombus, leading to cardioembolic stroke, especially in patients with atrial fibrillation (AF).1–3 Atrial fibrillation affects over 33 million people worldwide4 and causes atrial contraction to be ineffective, often causing blood stasis within the LAA.5 A non-invasive imaging test capable of evaluating the mechanical function of the LAA could potentially add valuable information toward stroke risk stratification leading to more precise application of anticoagulation therapy than currently available with CHA2DS2-VASc score6,7 and TEE alone.
The use of cardiac computed tomography (CT) to accurately detect existing thrombus8 and filling defects9–11 in the LAA has been well established; however, evaluating the blood flow and mechanical function of the LAA is more difficult. The LAA has very complex morphology that is highly variable between patients,12 causing difficulty in identifying the LAA orifice (the boundary between the LAA and the LA) with high reproducibility.13 The emergence of LAA occlusion devices has led to the development of procedures for the analysis of LAA geometry and orifice from transesophageal echocardiography (TEE) or CT, as well as detection of blood flow velocity through the orifice.14
Estimates of blood flow velocity through the LAA ostium can be directly made with TEE, and risk of stroke has been shown to weakly correlate with decreasing velocity15,16; however, LAA ejection fraction has poor correlation with the LAA velocity measure on TEE17. TEE also has limitations when compared to CT for estimating the morphology of the LAA. Clinically, TEE is a semi-invasive, uncomfortable procedure that typically requires an anesthetic. Analytically, the complex geometry of the LAA is incompletely characterized by single 2D views seen on TEE; the orientation of the views is limited by the restricted views available from the esophagus. Inconsistencies between morphological measurements made by CT and TEE in the same patient are well documented.18–21
Modern wide detector CT scanners with fast gantry rotation are capable of imaging functional dynamics of the LAA with cine CT; however, quantitative functional evaluation of the LAA ejection fraction (LAAEF), total LAA volume vs. time, and rate of change of volume per unit time) remains an open problem, primarily due to the lack of a standardized and reproducible procedure for defining the LAA ostium12. This is complicated further by the lack of an exact anatomical ostium on the LAA (Figure 1). Given the lobed, tapered, and/or fractal-like structure of the LAA22, a small change in one of the largest sections of the LAA, such as the ostium, can lead to radically different sizes of observed ostium and overall LAA volume. A definition of the LAA ostium that is consistent across all phases of the heart cycle, even in patients with radically different LAA anatomy, is required to reliably derive quantitative LAA functional information such as LAAEF.
Figure 1:
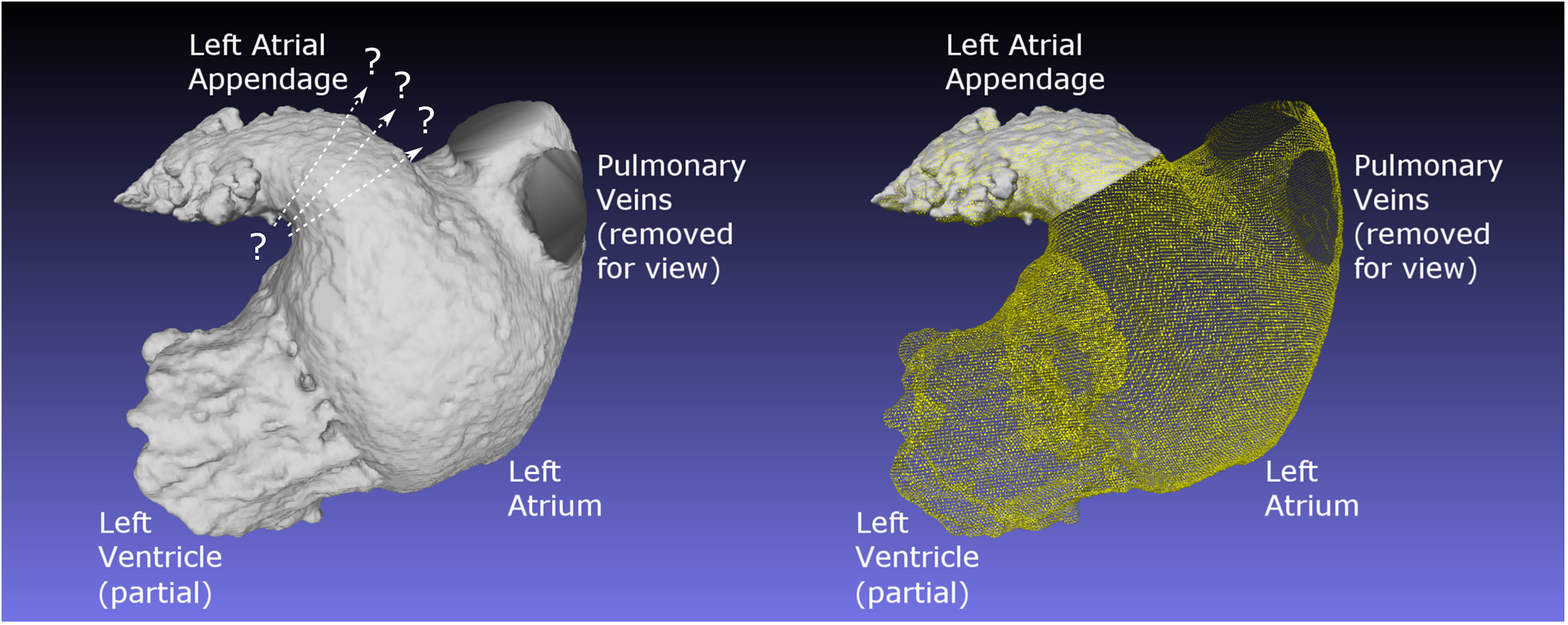
Image emphasizing the difficulty of making a precise and repeatable plane of separation between the LA and the LAA. On the left is a portion of the blood pool from the left side of the heart of Patient 2, with regions as noted. No clear separation of LA from LAA is notable. On the right, the same blood pool is shown, with the procedurally determined ostium made clear.
The purpose of this study was to evaluate the intra- and inter-observer reproducibility of a procedure for the definition of the LAA ostium in 3D CT volumes. Our focus is on the reproducibility of the definition of the LAA ostium – hence we test the intra- and inter-observer variability in the computed LAA volumes in 25 subjects with full 4D CT acquisitions including the LAA.
Methods
Because of the sensitive nature of the data collected for this study, requests to access the dataset from qualified researchers trained in human subject confidentiality protocols may be sent to the Cardiovascular Imaging Lab (CViL) at emcveigh@ucsd.edu.
Subjects
This multi-center, cross-sectional study was approved by the institutional review boards (IRBs) of all participating institutions (Radiology and Imaging Sciences, NIH Clinical Center; National Heart, Lung, and Blood Institute; and the Department of Radiology at the University of California, San Diego). Subject consent was not required for this retrospective study. Subjects scanned between 2012 and 2017 with full R-wave to R-wave (RR) cine capture were considered for inclusion. All subjects included in this study were referred to cardiac CT for either coronary artery imaging (28%) or TAVI planning (72%). Retrospectively gated cardiac cine CT was performed in all subjects.
Inclusion criteria were as follows: (1) the LAA was fully visible in the field of view in all time frames; (2) intravascular contrast was well timed and mixed correctly for LAA imaging (contrast mixing was present and did not significantly change over the course of the scan); (3) signal to noise ratio (SNR) in the LA was > 5; (4) a minimum of 9 cardiac phases were reconstructed, over a minimum of 90% of the cardiac cycle; 31 patients met all four criteria, from which 25 patients were selected to be included in the study. Two patients failed, due to unusual vasculature geometry precluding a simple rectangular volumetric region of interest (ROI) around the LAA.
Seven patients had 20 time frames reconstructed per heart cycle, while 17 patients had 10 time frames, and 1 patient had 9. A subset of the time frames that were separated in time by at least 360 degrees of gantry rotation were considered for the purposes of computing the ICC metrics, and for performing ANOVA. This was done to avoid temporal correlations between time frames due to raw data sharing. All time frames were used in creating volume vs. time curves, and for initially determining the minimum and maximum volume time frames. Minimum and maximum time frames were selected by determining the volume of all time frames of every patient and selecting the time frames corresponding to the most frequently minimum and maximum volumes as seen across all 9 observations in each patient. The relatively small number of patients used in this study was due to the limited number of available datasets of the LAA over a full heart cycle, as well as the amount of time needed to fully process each patient 9 times (9×25=225 segmentations).
The mean LA SNR of the patient datasets was 13.2 ± 2.8. Matrix size was 512×512 for all datasets, and slice thickness was 0.53mm ± 0.06. Mean X and Y pixel spacing was 0.417×0.417mm ± 0.054. Hounsfield Unit (HU) values were calculated after multi-planar reconstruction (MPR) processing so that the HU values of the LAA ostium were used, and not of the LA or the more distal regions of the LAA.
Initial subject data is summarized in Table 1.
Table 1. Initial Patient Data.
The characteristics of the patients and scans used in the study.
| Patient | Manufacturer | Machine | Sex | Age (yrs) | Dose Index (mGy) | AF? |
|---|---|---|---|---|---|---|
| Patient 01 | Siemens | SOMATOM Force | M | 62 | 32.05 | no |
| Patient 02 | Siemens | SOMATOM Force | F | 53 | 20.97 | no |
| Patient 03 | Siemens | SOMATOM Force | M | 55 | 19.94 | no |
| Patient 04 | Siemens | SOMATOM Force | F | 60 | 21.90 | no |
| Patient 05 | Siemens | SOMATOM Force | M | 71 | 10.97 | no |
| Patient 06 | Siemens | SOMATOM Force | M | 53 | 16.45 | no |
| Patient 07 | GE | Discovery CT750 HD | M | 91 | 67 | yes |
| Patient 08 | GE | Discovery CT750 HD | F | 79 | 69.7 | no |
| Patient 09 | GE | Discovery CT750 HD | M | 85 | 21.9 | no |
| Patient 10 | GE | Discovery CT750 HD | M | 88 | 72.2 | no |
| Patient 11 | Toshiba | Aquillion ONE | M | 80 | 26.7 | no |
| Patient 12 | GE | Discovery CT750 HD | F | 84 | 73 | no |
| Patient 13 | Toshiba | Aquillion ONE | M | 92 | 21.2 | yes |
| Patient 14 | Toshiba | Aquillion ONE | M | 67 | 70.1 | yes |
| Patient 15 | Toshiba | Aquillion ONE | F | 90 | 17.9 | no |
| Patient 16 | Toshiba | Aquillion ONE | M | 82 | 14.7 | yes |
| Patient 17 | Toshiba | Aquillion ONE | F | 78 | 27.5 | yes |
| Patient 18 | Toshiba | Aquillion ONE | F | 90 | 26.4 | no |
| Patient 19 | Toshiba | Aquillion ONE | M | 85 | 39.1 | no |
| Patient 20 | Toshiba | Aquillion ONE | M | 81 | 19.3 | no |
| Patient 21 | Toshiba | Aquillion ONE | F | 89 | 29.8 | yes |
| Patient 22 | Toshiba | Aquillion ONE | M | 71 | 31.6 | no |
| Patient 23 | GE | Revolution CT | M | 73 | 23.80 | no |
| (failed) | Toshiba | Aquillion ONE | M | 79 | 10.30 | no |
| Mean: | - | - | - | 75.51 | 27.59 | - |
AF: patient scanned while in atrial fibrillation
Image analysis
The analysis procedure consisted of four steps: (1) identification of the anatomical landmarks defining the plane of the LAA orifice; (2) creating the MPR of the CT data aligned with this plane; (3) segmentation of the LAA from the MPR images using a semiautomated active contour region-growing procedure; and (4) calculation of relevant functional parameters from these segmentations.
LAA Ostium Segmentation
The 3D CT data was re-oriented spatially using multi-planar reconstruction (MPR), which was conducted independently for each time frame, observer, and trial using the OsiriX software (version 5.5.2).23 The key steps of the MPR procedure are depicted in Figure 2; a detailed description is given in the Appendix.
Figure 2.
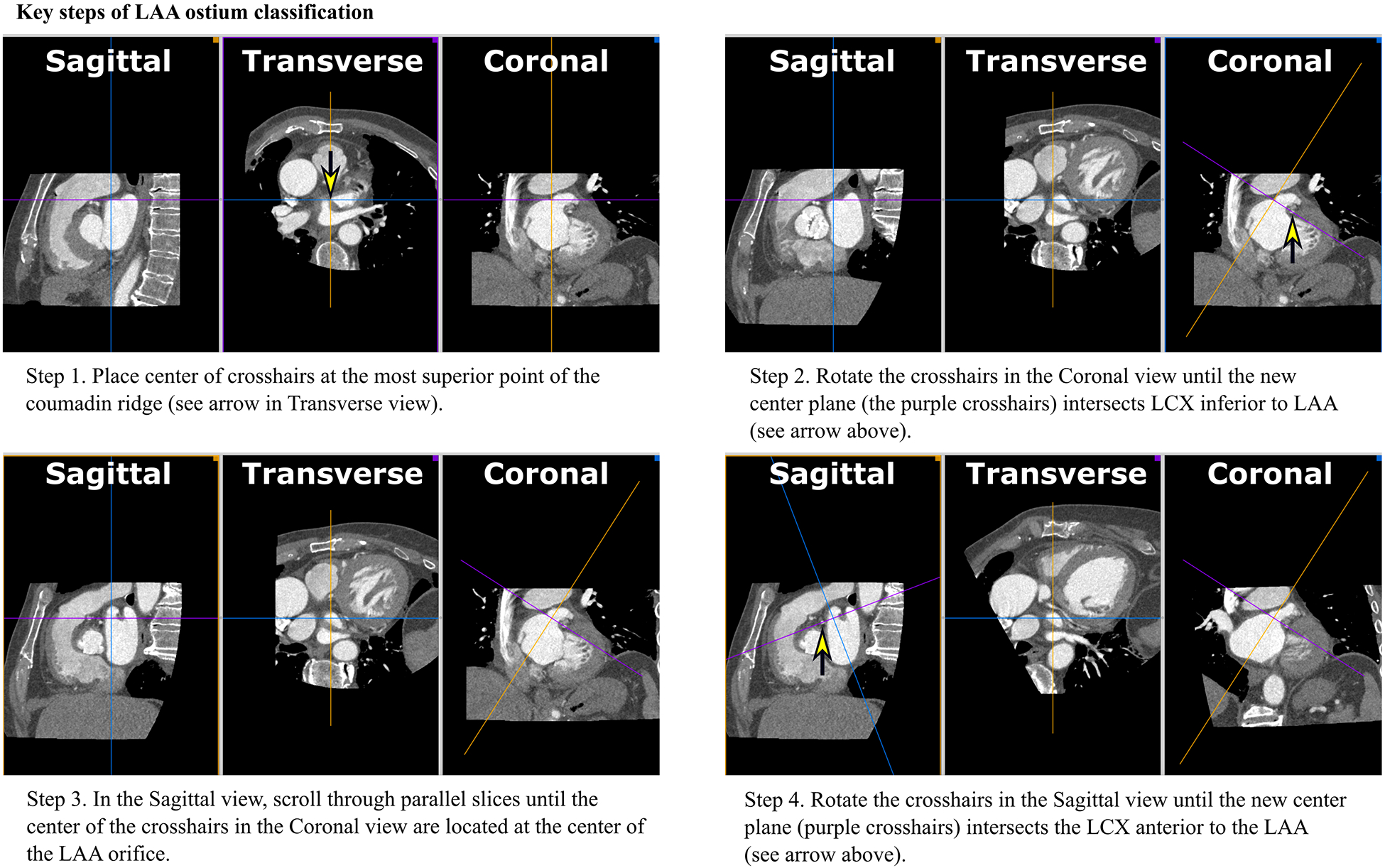
Key steps of methodological LAA orifice classification using multi-planar reconstruction (MPR)
The crosshairs of Figure 2. show the intersection of two spatial planes which lie perpendicular to the plane of the displayed image.
First, the center of the crosshairs, when viewed in the Transverse plane, was placed on the most superior portion of the Coumadin ridge, identified as a visual separation between the left atrium and LAA (see arrow in Fig 2.a.). This separation merged and separated further when moving back and forth through the Transverse planes. Then, in the Coronal view, the crosshairs were rotated until the purple crosshair intersected the left circumflex artery (LCX) inferior to the LA (Fig 2.b.). Next, as shown in Fig 2.c., the operator paged back and forth through the Sagittal planes while viewing the position of the cursor in the Coronal plane; the operator positioned the center of the crosshairs at the center of the LAA orifice in the Coronal plane. Finally, in the Sagittal view, the crosshairs were rotated so that the purple plane (see Fig 2.d.) intersected the LCX in the Sagittal view. Together, these reorientations formed a plane that contained the LAA ostium, now displayed in the Transverse view (red contour in Fig 2.d.). A stack of parallel planes from this location to the most distal extend of the LAA were created in OsiriX. The final inter-plane spacing was calculated automatically by OsiriX on a per-patient basis, resulting in a mean slice thickness of 0.207mm ± 0.027 for all patient MPRs; this was a significant oversampling of the original resolution, ensuring that details were not lost due to undersampling. This resliced volume was subsequently analyzed to measure LAA volume vs. time.
To begin segmenting the LAA blood volume, a ROI was drawn in a slice that was centrally located within the LAA blood pool and surrounding tissue using OsiriX. A histogram was generated for all pixels within this ROI, and the HU value which separated the LAA blood from the surrounding tissue wall was chosen as the threshold for downstream segmentation (as shown in Figure 3). Values were rounded to the nearest tens place whenever an exact value was not distinguishable; however, exact values for thresholding were not critical, as thresholding values were maintained constant for each patient. This was to ensure that the focus of the study remained on the MPR method described here. Mean threshold value was 277.58 ± 109.01. Minimum threshold was 125, and maximum was 700.
Figure 3:
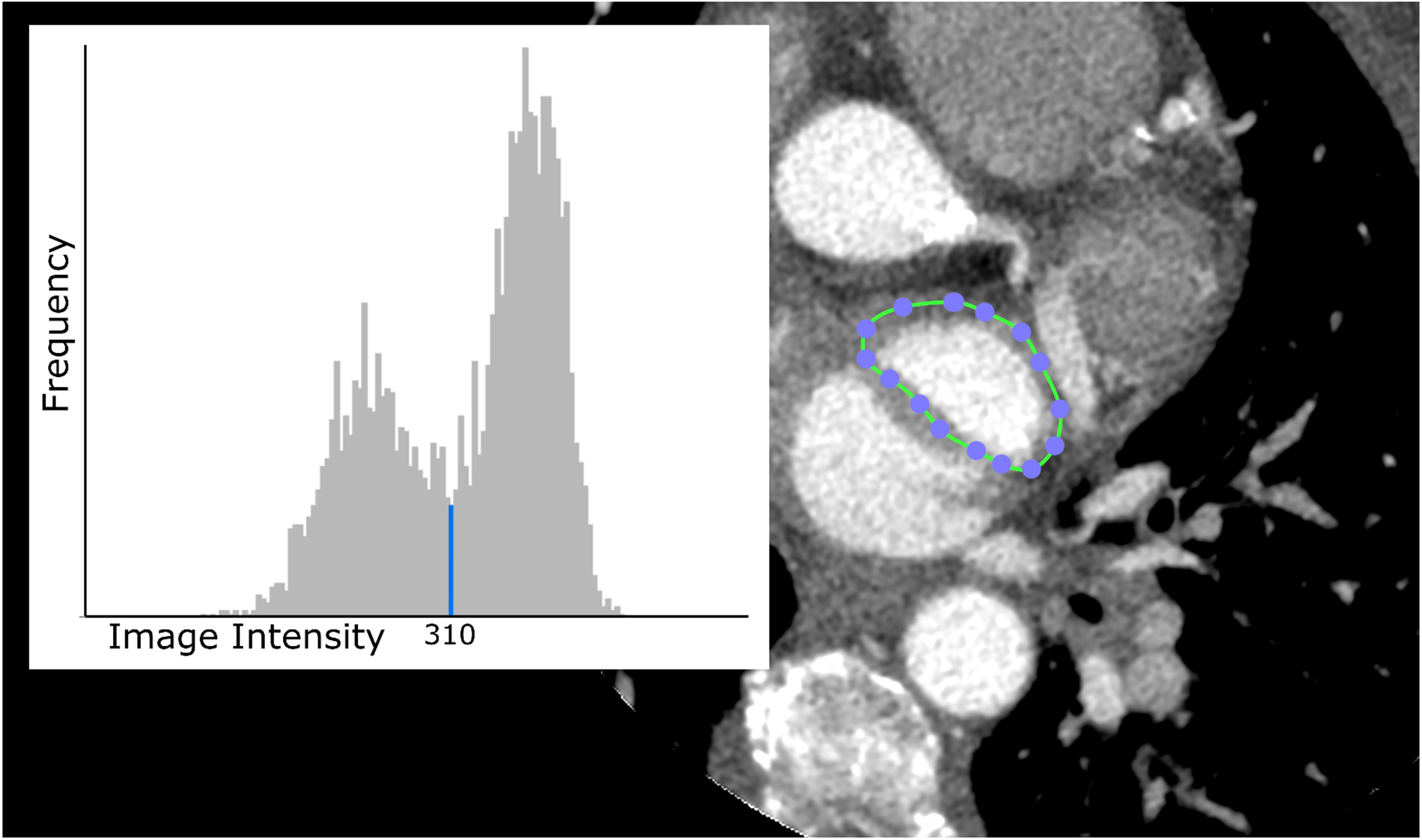
Screenshot excerpt from the step in the procedure designed to extract a threshold value for use in segmenting the blood volume of the LAA to form a mesh volume. A region of interest (ROI) is drawn around the LAA with the closed area ROI tool including blood and surrounding tissue; the image intensity values within the ROI are displayed in a histogram. The histogram shows a local minimum corresponding to the values primarily around the edge of the bright LAA region. The values above the threshold were chosen to segment the LAA blood volume. (Image Intensity is in Hounsfield Units, the Frequency is relative number of voxels.)
Segmentation of the LAA blood was performed using the active contour segmentation mode of ITK-Snap (version 3.6.0)24 with the previously determined threshold. One to 3 initialization spheres (referred to as “bubbles”24) with an approximate radius of 5 mm were added along the long axis of the LAA. Smaller bubbles (typically around 2mm) were used to ensure segmentation of distal regions and smaller trabeculations within the LAA. Starting from this initialization, active contour segmentation proceeded until the LAA was filled, based on visual inspection. The boundary of the resulting segmentation was saved as a mesh in STL format. The full listing of mesh-growing parameters used by ITK-Snap for these segmentations are contained within Appendix B. This step and all subsequent steps were performed by a single operator, or methodologically within ITK-Snap, removing the independence of the data with respect to all observers and trials from this point forward, ensuring that the variation of the results remained focused on the method used to define the plane of the ostium outlined in Section “LAA Ostium Segmentation”.
The resulting STL files from the segmentations were loaded into Meshlab to correct for any errors in the meshes.25 In a small number of volumes, topological errors (vertex duplicates, erroneously joined structures, etc.) in the mesh were easily resolved through manual editing in Meshlab. In roughly 20% of all volumes, where the segmentation “leaked” into neighboring vessels (e.g., the left superior pulmonary vein), the mesh was manually pruned to include only the LAA. Editing was minimal, and was performed solely by one observer, to ensure consistency in LAA wall segmentation because the purpose of this study was to show the consistency of the ostium segmentation. Once the meshes were made contiguous, the volume contained within the LAA mesh was computed.
Statistical Analysis and Reproducibility
The goal of this study was to quantify the reproducibility and precision of this landmark-based method of LAA ostium classification. Intra-observational data were collected to determine the precision of the results when repeated analysis was performed by the same observer, and inter-observational data were collected to establish the precision of results across the different observers. The observers were two bioengineering undergraduate students (KL, CJ) and one graduate student (AS), none of whom had formal clinical medical training. Each of the three observers measured the LAA volume three times across all patients and cardiac time frames. Patients were not analyzed in any specified order, and the observers were blinded to the identity of the cases during the analysis. Direct results of the volume analysis included maximum and minimum volumes, standard deviations around the observer, group mean, and LAAEF in all patients.
For statistical analysis, the data were organized into an n × k table, where the rows were the n=225 individual volume estimates (23 patients × 9 time frames) and k=9 measurements (3 observers × 3 measurements each). Note that “rows” and “columns”, as mentioned below, are the terms of the ICC function. In this analysis, the observers formed the “rows,” and the volume measurements formed the “columns.” The 2 failed patients were unable to be analyzed and were not included here. Intraclass correlation coefficients (ICCs) were obtained using the two-way random effect, absolute agreement, single measurement equation below:
| (1) |
MSR = mean square for ‘rows’ (the variance of the mean of all measurements of each volume, multiplied by the number of observers); MSE = mean square error (also referred to as “residual mean square”); MSC = mean square for ‘columns’ (the variance of the mean of each set of volume measurements, multiplied by the total number of volumes); n = number of volumes; k = number of observers or measurements (for interobserver and intraobserver, respectively).
The ICC equation selected was chosen in accordance with Koo and Mae.26 Two-way random effect was chosen to reflect the fact that although the same observers were used throughout the study, the observers were random with respect to the data obtained; that is, the results were not intended to reflect the observers as individuals, but as a sample of potential observers. Absolute agreement was chosen to determine how precise the method was, but not its accuracy, as ground truth data could not be obtained. Single measurement was selected as all observers took a single sample at each time frame of concern, within each trial. ICC values were calculated at a 95% confidence interval. Using the recommended values provided by Koo and Mae26 for a 95% confidence interval (poor, moderate, good, and excellent, corresponding respectively to ICC values of less than 0.5, 0.5 to 0.75, 0.75 to 0.9, and above 0.9), values over 0.9 convey excellent correspondence.
ICCs were calculated using the “icc” function of the “irr” package in R27,28, and verified using the Real Statistics Resource Pack for Excel29.
Results
Initial data of all patients is tabulated in Table 1.
Mean HU of the LAA blood at the ostium slice of each patient’s MPR set was 474.09 ± 126.95. For each image slice, the HU values were gathered by selecting the largest circular or ellipsoidal ROI possible that was fully enclosed by the LAA (ROI mean area of 157.09 mm2 ± 101.62), and extracting the numerical data using ImageJ30,31. HU analysis was limited to each patient MPR dataset by observer 1, trial 1 only.
Minimum and maximum values of the LAA volumes measured using this technique for each patient are shown in Table. 2.
Table 2: Range of observed LAA volume data.
Shown categories are the maximum volumes of each patient, the minimum volumes, and the left atrial appendage ejection fraction (LAAEF). LAA volumes are expressed as the mean ± standard deviation (SD) over all 9 observations (3 observers and 3 repeated analyses) and average LAAEF percentage (%).
| Patient | Max Volume (mL) ± SD | Min Volume (mL) ± SD | LAAEF* (%) ± SD |
|---|---|---|---|
| Patient 01 | 7.99 ± 0.12 | 2.71 ± 0.09 | 66.07 ± 0.95 |
| Patient 02 | 10.01 ± 0.43 | 1.75 ± 0.16 | 82.53 ± 1.02 |
| Patient 03 | 17.85 ± 0.32 | 7.53 ± 0.19 | 57.81 ± 0.7 |
| Patient 04 | 5.81 ± 0.17 | 1.97 ± 0.16 | 65.85 ± 2.9 |
| Patient 05 | 8.78 ± 0.14 | 4.16 ± 0.31 | 52.3 ± 3.64 |
| Patient 06 | 10.97 ± 0.24 | 3.74 ± 0.12 | 65.9 ± 0.6 |
| Patient 07 | 10.16 ± 0.15 | 7.38 ± 0.24 | 27.15 ± 2.71 |
| Patient 08 | 13.14 ± 1.12 | 5.04 ± 0.27 | 61.55 ± 2.06 |
| Patient 09 | 5.31 ± 0.11 | 2.82 ± 0.19 | 46.7 ± 3.46 |
| Patient 10 | 29.7 ± 0.73 | 15.51 ± 0.56 | 47.64 ± 2.69 |
| Patient 11 | 12.06 ± 0.88 | 7.21 ± 0.51 | 39.83 ± 4.11 |
| Patient 12 | 12.72 ± 0.3 | 7.97 ± 0.49 | 37.13 ± 3.16 |
| Patient 13 | 22.65 ± 1.05 | 18.4 ± 1.2 | 17.92 ± 5.37 |
| Patient 14 | 11.78 ± 0.91 | 6.65 ± 0.28 | 43.22 ± 3.95 |
| Patient 15 | 12.25 ± 0.18 | 6.31 ± 0.16 | 48.44 ± 1.74 |
| Patient 16 | 10.14 ± 0.3 | 8.02 ± 0.29 | 20.85 ± 1.67 |
| Patient 17 | 9.49 ± 0.58 | 6.25 ± 0.47 | 33.92 ± 3.01 |
| Patient 18 | 5.93 ± 0.34 | 3.3 ± 0.42 | 43.15 ± 7.77 |
| Patient 19 | 8.22 ± 0.22 | 3.42 ± 0.33 | 58.11 ± 3.72 |
| Patient 20 | 6.93 ± 0.49 | 2.29 ± 0.05 | 66.8 ± 2.51 |
| Patient 21 | 11.49 ± 0.77 | 8.77 ± 0.55 | 23.56 ± 2.07 |
| Patient 22 | 11.44 ± 0.69 | 5.15 ± 0.63 | 54.7 ± 3.69 |
| Patient 23 | 7.24 ± 0.11 | 2.42 ± 0.2 | 66.46 ± 2.38 |
Notes:
LAA Ejection Fraction (defined as (1 - Volmin/Volmax)*100%)
SD: Standard deviation
The mean and standard deviation of the maximum volume (averaged across all observations) of each patient in Table 2 were 10.31 mL ± 5.25, while the mean of each averaged minimum volume was 5.02 mL ± 4.07, and the mean LAAEF was 45.69 % ± 16.65; The high standard deviations were due to the wide range of volumes observed, so it is critical to emphasize the very unique morphology of each patient’s LAA in clinical practice rather than the statistical average.
Example volume vs. time data from 3 patients who had very different LAA size and LAAEF are shown in Figure 4, showing the wide range of values in the data.
Figure 4:
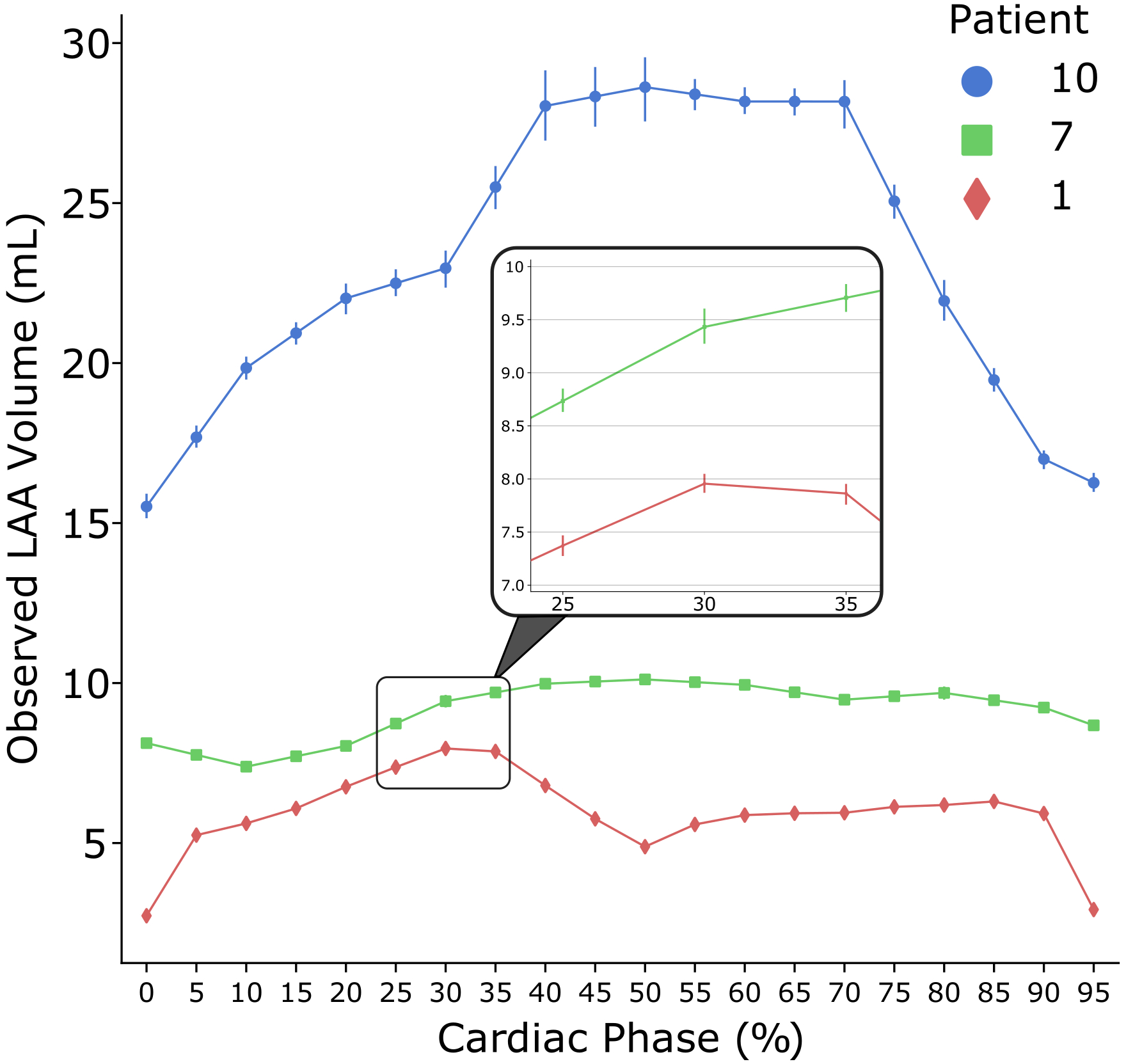
Graphical representation of the left atrial appendage (LAA) volume vs. time over the heart cycle for three different patients. The data points represent the mean value across all nine observations (three repeated analyses from each of the three observers); the bars represent the standard deviation (SD) of the 9 measurements around the mean. The three patients displayed were selected to show the variability of LAA size and ejection fraction.
Note: The SD bars for Patients 1 and 7 are too small to be visible as shown and are depicted in greater detail within the zoomed-in window.
High intraclass correlation coefficients (ICC) were found for intraobserver comparisons (ICCs 0.989, 0.989, and 0.995, with lower/upper bounds .987/.991, .987/991, and .994/.996, for observers 1, 2, and 3, respectively) and interobserver comparisons (ICCs 0.984, 0.990, 0.988, with lower/upper bounds .981/.987, .987/.992, and .985/.990, for trials 1, 2, and 3, respectively) of LAA volume analysis. The ICC of all observations, across all observers, was 0.988 (lower/upper bounds .986/.990).
An alternative quantitative representation of the high precision and reproducibility of this technique is shown in Figure. 5 in which 6 example correlation plots are shown (out of 36 possible), comparing the measurements of LAA volume from the following subset of measurements:
Figure 5:
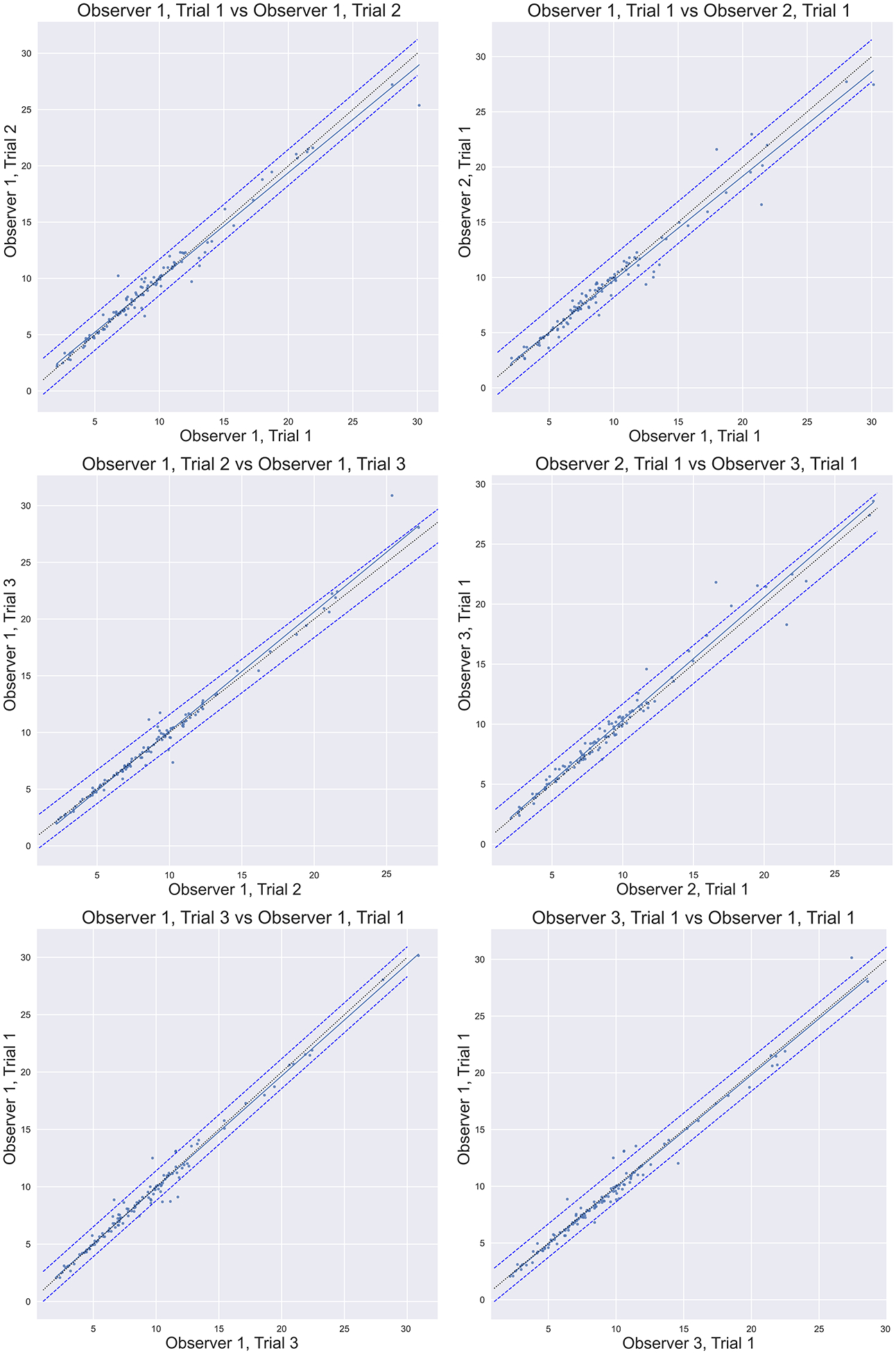
Correlation plots comparing the measurements of LAA volume from all 3 of the observer 1 trials (intraobserver correlation), and comparing the first trials of all observers (interobserver correlation) for a subset of the comparisons. In each plot, over 230 measurements of LAA volume in cm3 are depicted with a linear trendline (solid line). Error bars depict the 95% confidence interval. Most of the 36 possible permutations of the data to be compared have been omitted for simplicity but display similar data to the permutations shown. Line of identity is shown as a thin dotted line.
Fig 5.a. (Observer 1, trial 1 vs. Observer 1, trial 2), Fig 5.b. (Observer 1, trial 1 vs. Observer 2, trial 1), Fig 5.c. (Observer 1, trial 2 vs. Observer 1, trial 3), Fig 5.d. Observer 2, trial 1 vs. Observer 3, trial 1), Fig 5.e. (Observer 1, trial 1 vs. Observer 1, trial 3), Fig 5.f. Observer 1, trial 1 vs. Observer 3, trial 1).
In each plot, over 600 measurements of LAA volume in cm3 are depicted, and a linear trendline is displayed (solid line) with the parameters of the fit given in the respective figure panels. Error bars depict the 95% confidence interval of the data correlation (dashed lines), calculated as the standard deviation of the arithmetic difference between the data, multiplied by 1.96, and applied both above and below the trendline. The line of identity is shown as a dotted line.
Figure 6. demonstrates the coefficient of variation (CV) of the LAA volumes.
Figure 6.
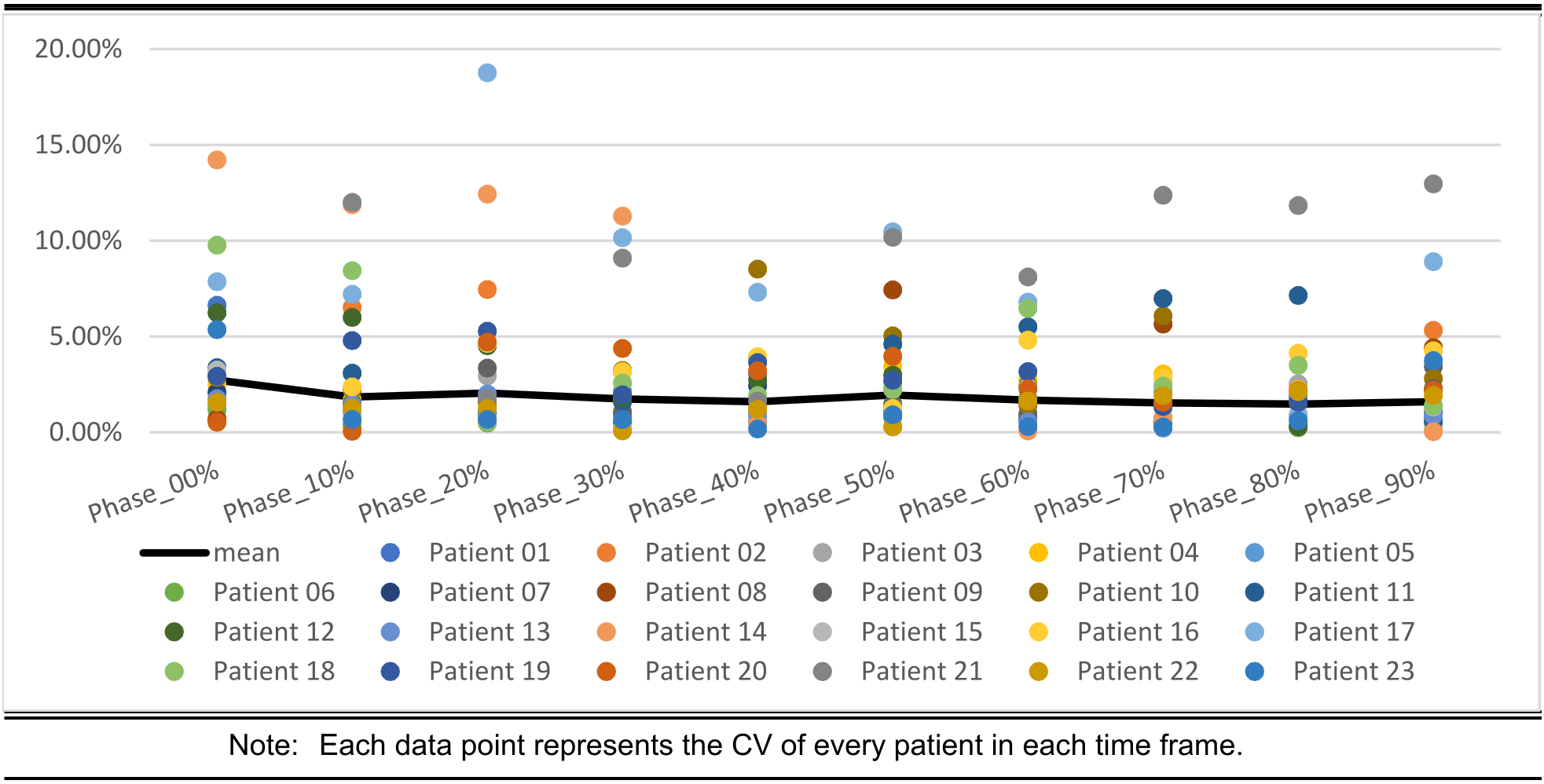
Coefficient of variation (CV) of the LAA volume for each of the respective phases of the heart, and across all patients
Discussion
In this study, a total of 2736 LAA ostia were segmented and LAA volume measurements made, from which 1035 LAA ostia were used for ICC analysis (23 patients, 5 time frames, 9 measurements per time frame). The purpose was to test the reproducibility of an algorithm for defining the division of the LAA from the LA. In order to isolate our examination of the reproducibility of this process, we used a very simple volume segmentation of the LAA, and the same segmentation parameters were used for the 9 different ostial surfaces created for each timeframe, defined by the process described in this paper. The volumes and LAAEF values spanned a large dynamic range. Figure 5 demonstrates this range and the high confidence in the reproducibility of the technique between operators and between trials of the same operator.
In order to ensure that each volume had independent noise we analyzed time frames that were separated by a time that was greater than a 360-degree rotation of the scanner gantry. In particular, for each patient we used 5 LAA volumes from nonconsecutive time frames as the datapoints for ICC and ANOVA analysis for each patient (specifically, phases 0%, 20%, 40%, 60%, and 80% of each patient’s R-R interval). Dose modulation scans were not included in order to isolate the effect of cardiac phase from changing SNR.
Volumes were calculated inside meshes, instead of voxel-counting the original segmentations, to allow for ease of manual post-processing, such as removal of spurious components outside the LAA that the ITK-snap segmentation included.
This technique may prove extremely helpful not only for characterizing the LAA ejection fraction as an assessment of risk of thrombosis, but also for planning LAA isolation procedures such as occlusion device implantations. Removing the variability of the data obtained by different clinical observers would increase the reproducibility of sizing the LAA compared to the techniques used currently, such as 2D TEE, or 3D CT without a precisely defined segmentation procedure.
The method does suffer from the need for human supervised segmentation, which is a disadvantage. However, the process of reproducibly defining the plane which defines the ostium requires about 30 seconds for an experienced user. Automated segmentation would be a logical progression of this technique, including the segmentation of the LAA and LA blood pool. It is conceivable that a neural network could be used to produce a binary map of voxels “inside” vs. “outside” the LAA; however, that neural network will need accurate data from which to learn. The technique outlined herein could provide those training data sets.
Limitations
Some LAA patients analyzed required manual post-processing to eliminate additional volumes created by “leaks” in the segmentation. In many cases, the process used to create the LAA meshes included adjacent vessels (most often the pulmonary veins when they were “fused” with the LAA and left circumflex artery). However, in these cases, it was simple to manually correct the segmentation to test our hypothesis.
The primary reason for inconsistencies in the result of the technique was the selection of the wrong landmark vessel in some of the time frames, if the LCX became hidden by artifacts, or the coronary sinus (or another vessel) was incorrectly identified as the LCX. Non-optimal contrast agent timing may have enhanced the error, since arteries of reduced image intensity sometime resemble the coronary sinus or other vessels.
The two patients that were considered “failures” produced ostia that partly bisected the LAA and did not form a boundary between the LA and LAA. The data from these patients was therefore not included in the analysis. Using a curved MPR plane could possibly compensate for these failures, but this was not included in this study because we desired to keep the process as simple as possible. Patients with highly unusual morphology should likely be assessed individually.
From our data the predictor of failure is an irregular trajectory of the proximal circumflex artery. The presence of AF and its impact on image quality was not studied, although we did not find evidence suggesting that AF would negatively impact LAA image acquisition or this technique. Correlations between cardiac rhythm and metrics such as LAA EF have been noted15, but were not evaluated in this study.
Accuracy of this procedure was not quantified, as no ground truth existed for the data analyzed; however, for LAAEF calculations and other physiological biomarkers computed for individual patients, it is most important that a reproducible boundary is available at the key time frames, which has been provided by this method.
Conclusions
Highly reproducible segmentation of the LAA ostium was achieved using a simple ordered landmark identification and 3D multi-planar reorientation method to construct a plane dividing the LAA from the LA. This algorithm will allow precise evaluation of LAA function through the accurate calculation of LAA ejection fraction in the large majority of patients.
Supplementary Material
Potential Clinical Impact.
The ability to measure the function of the left atrium (LA) and left atrial appendage (LAA) may be critical for improving the precision of evaluating stroke risk, especially in patients with atrial fibrillation. Current methods such as estimating the peak velocity of blood through the LAA orifice with TEE have only weak correlations with LAA ejection fraction and patient outcomes. Cine CT provides superb spatial resolution for calculating LAA ejection fraction; however, a current limitation is our inability to identify a reproducible boundary between the LAA and the LA over the heart cycle. In this paper, we describe a procedure for defining the LA/LAA boundary using anatomical landmarks which proves to be highly reproducible between observers and trials from images obtained from multiple CT scanners. This simple procedure coupled with LAA blood segmentation would seem to provide the basis for gathering clinical data on LAA function from many patients imaged on different systems.
Acknowledgements
We thank David A. Bluemke MD, PhD, Veit Sandford MD for collaborating in collecting the Siemens Force data used in this study.
Sources of Funding
None.
Abbreviations:
- CT
computed tomography
- LA
left atrium
- LAA
left atrial appendage
- TEE
transesophageal echocardiography
- TAVI
transcatheter aortic valve implantation
- AF
atrial fibrillation
- LAAEF
left atrial appendage ejection fraction
- SNR
signal-to-noise ratio
- MPR
multi-planar reconstruction
- LCX
left circumflex artery
- SD
standard deviation
- ICC
intraclass correlation coefficient
- ANOVA
Analysis of variance
- ROI
Region of interest
- CTDIvol
Computed Tomography Dose Index (volume)
Footnotes
Disclosures:
Dr. McVeigh is a co-founder and shareholder of MRI Interventions Inc.
Dr. McVeigh receives research funding from: GE Healthcare, Pacesetter Inc., Tendyne Holdings Inc.
References
- 1.Al-Saady NM, Obel OA, Camm AJ. Left atrial appendage: structure, function, and role in thromboembolism. Heart. 1999; 82:547–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yaghi S, Song C, Gray WA, Furie KL, Elkind MS V, Kamel H. Left Atrial Appendage Function and Stroke Risk. Stroke. 2015; 46:3554–3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beigel R, Wunderlich NC, Ho SY, Arsanjani R, Siegel RJ. The left atrial appendage: Anatomy, function, and noninvasive evaluation. JACC Cardiovasc Imaging. 2014; 7:1251–1265. [DOI] [PubMed] [Google Scholar]
- 4.Chugh SS, Havmoeller R, Narayanan K, Singh D, Rienstra M, Benjamin EJ, Gillum RF, Kim YH, McAnulty JH, Zheng ZJ, et al. Worldwide epidemiology of atrial fibrillation: A global burden of disease 2010 study. Circulation. 2014; 129:837–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Markl M, Lee DC, Furiasse N, Carr M, Foucar C, Ng J, Carr J, Goldberger JJ. Left Atrial and Left Atrial Appendage 4 D Blood Flow Dynamics in Atrial Fibrillation. Circ Cardiovasc Imaging. 2016; 9:e004984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lip GYH, Nieuwlaat R, Pisters R, Lane DA, Crijns HJGM. Refining clinical risk stratification for predicting stroke and thromboembolism in atrial fibrillation using a novel risk factor-based approach: The Euro Heart Survey on atrial fibrillation. Chest. 2010; 137:263–272. [DOI] [PubMed] [Google Scholar]
- 7.Lee JM, Kim JB, Uhm JS, Pak HN, Lee MH, Joung B. Additional value of left atrial appendage geometry and hemodynamics when considering anticoagulation strategy in patients with atrial fibrillation with low CHA2DS2-VASc scores. Heart Rhythm. 2017; 14:1297–1301. [DOI] [PubMed] [Google Scholar]
- 8.Romero J, Husain SA, Kelesidis I, Sanz J, Medina HM, Garcia MJ. Detection of left atrial appendage thrombus by cardiac computed tomography in patients with atrial fibrillation: A meta-analysis. Circ Cardiovasc Imaging. 2013; 6:185–194. [DOI] [PubMed] [Google Scholar]
- 9.Kim YY, Klein AL, Halliburton SS, Popovic ZB, Kuzmiak SA, Sola S, Garcia MJ, Schoenhagen P, Natale A, Desai MY. Left atrial appendage filling defects identified by multidetector computed tomography in patients undergoing radiofrequency pulmonary vein antral isolation: A comparison with transesophageal echocardiography. Am Heart J. 2007; 154:1199–1205. [DOI] [PubMed] [Google Scholar]
- 10.Yasuoka R, Kurita T, Kotake Y, Akaiwa Y, Hashiguchi N, Motoki K, Yamamoto H, Kobuke K, Iwanaga Y, Hirano Y, et al. A novel method to estimate blood flow velocity in the left atrial appendage using enhanced computed tomography: role of Hounsfield unit density ratio at two distinct points within the left atrial appendage. Heart Vessels. 2017; 32:893–901. [DOI] [PubMed] [Google Scholar]
- 11.Sawit ST, Garcia-Alvarez A, Suri B, Gaztanaga J, Fernandez-Friera L, Mirelis JG, D’Anca M, Fuster V, Sanz J, Garcia MJ. Usefulness of cardiac computed tomographic delayed contrast enhancement of the left atrial appendage before pulmonary vein ablation. Am J Cardiol 2012; 109:677–684. [DOI] [PubMed] [Google Scholar]
- 12.Wang Y, Di Biase L, Horton RP, Nguyen T, Morhanty P, Natale A. Left atrial appendage studied by computed tomography to help planning for appendage closure device placement. J Cardiovasc Electrophysiol. 2010; 21:973–982. [DOI] [PubMed] [Google Scholar]
- 13.López-Mínguez JR, González-Fernández R, Fernández-Vegas C, Millán-Nuñez V, Fuentes-Cañamero ME, Nogales-Asensio JM, Doncel-Vecino J, Elduayen-Gragera J, Ho SY, Sánchez-Quintana D. Anatomical classification of left atrial appendages in specimens applicable to CT imaging techniques for implantation of amplatzer cardiac plug. J Cardiovasc Electrophysiol. 2014; 25:976–984. [DOI] [PubMed] [Google Scholar]
- 14.Agmon Y, Khandheria BK, Gentile F, Seward JB. Echocardiographic assessment of the left atrial appendage. J Am Coll Cardiol. 1999; 34:1867–1877. [DOI] [PubMed] [Google Scholar]
- 15.Pollick C, Taylor D. Assessment of Left Atrial Appendage Function by Transesophageal Echocardiography\rImplications for the Development of Thrombus. Circulation. 1991; 84:223–231. [DOI] [PubMed] [Google Scholar]
- 16.Fukushima K, Fukushima N, Kato K, Ejima K, Sato H, Fukushima K, Saito C, Hayashi K, Arai K, Manaka T, et al. Correlation between left atrial appendage morphology and flow velocity in patients with paroxysmal atrial fibrillation. Eur Heart J Cardiovasc Imaging. 2016; 17:59–66. [DOI] [PubMed] [Google Scholar]
- 17.Matsumoto Y, Morino Y, Kumagai A, Hozawa M, Nakamura M, Terayama Y, Tashiro A. Characteristics of Anatomy and Function of the Left Atrial Appendage and Their Relationships in Patients with Cardioembolic Stroke: A 3-Dimensional Transesophageal Echocardiography Study. J Stroke Cerebrovasc Dis. 2017; 26:470–479. [DOI] [PubMed] [Google Scholar]
- 18.Saw J, Fahmy P, Spencer R, Prakash R, McLaughlin P, Nicolaou S, Tsang M. Comparing Measurements of CT Angiography, TEE, and Fluoroscopy of the Left Atrial Appendage for Percutaneous Closure. J Cardiovasc Electrophysiol. 2016; 27:414–422. [DOI] [PubMed] [Google Scholar]
- 19.Wang DD, Eng M, Kupsky D, Myers E, Forbes M, Rahman M, Zaidan M, Parikh S, Dnp JW, Pantelic M, et al. Application of 3-Dimensional Computed Tomographic Image Guidance to WATCHMAN Implantation and Impact on Early Operator Learning Curve. JACC Cardiovasc Interv. 2016; 9:2329–2340. [DOI] [PubMed] [Google Scholar]
- 20.Xu B, Betancor J, Sato K, Harb S, Abdur Rehman K, Patel K, Kumar A, Cremer PC, Jaber W, Rodriguez LL, et al. Computed tomography measurement of the left atrial appendage for optimal sizing of the Watchman device. J Cardiovasc Comput Tomogr. 2017; 12:50–55. [DOI] [PubMed] [Google Scholar]
- 21.Rajwani A, Nelson AJ, Shirazi MG, Disney PJS, Teo KSL, Wong DTL, Young GD, Worthley SG. CT sizing for left atrial appendage closure is associated with favourable outcomes for procedural safety. Eur Heart J Cardiovasc Imaging. 2017; 18:1361–1368. [DOI] [PubMed] [Google Scholar]
- 22.Beutler DS, Gerkin RD, Loli AI. The Morphology of Left Atrial Appendage Lobes: A Novel Characteristic Naming Scheme Derived through Three-Dimensional Cardiac Computed Tomography. World J Cardiovasc Surg. 2009; 4:17–24. [Google Scholar]
- 23.Rosset A, Spadola L, Ratib O. OsiriX: An open-source software for navigating in multidimensional DICOM images. J Digit Imaging. 2004; 17:205–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yushkevich PA, Piven J, Hazlett HC, Smith RG, Ho S, Gee JC, Gerig G. User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. NeuroImage. 2006; 31:1116–1128. [DOI] [PubMed] [Google Scholar]
- 25.Cignoni P, Cignoni P, Callieri M, Callieri M, Corsini M, Corsini M, Dellepiane M, Dellepiane M, Ganovelli F, Ganovelli F, et al. MeshLab: an Open-Source Mesh Processing Tool. Computing. 2008; 1:129–136. [Google Scholar]
- 26.Koo TK, Li MY. A Guideline of Selecting and Reporting Intraclass Correlation Coefficients for Reliability Research. J Chiropr Med. 2016; 15:155–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.R Core Team. R: A Language and Environment for Statistical Computing. 2018. Available from: http://www.R-project.org/. Accessed 02 Aug 2019.
- 28.Gamer Matthias L J and IFPS. irr: Various Coefficients of Interrater Reliability and Agreement. 2012. Available from: https://cran.r-project.org/package=irr. Accessed 12 Nov. 2019.
- 29.Zaiontz C. Real Statistics Using Excel. 2018. Available from: www.real-statistics.com. Accessed 02 Aug 2019.
- 30.Schindelin J, Rueden CT, Hiner MC, Eliceiri KW. The ImageJ ecosystem: An open platform for biomedical image analysis. Mol Reprod Dev. 2015; 82:518–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012; 9:671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


