Figure 1. Design of isoTarget and Its Application to Studying the Functional Diversity of Splicing Isoforms.
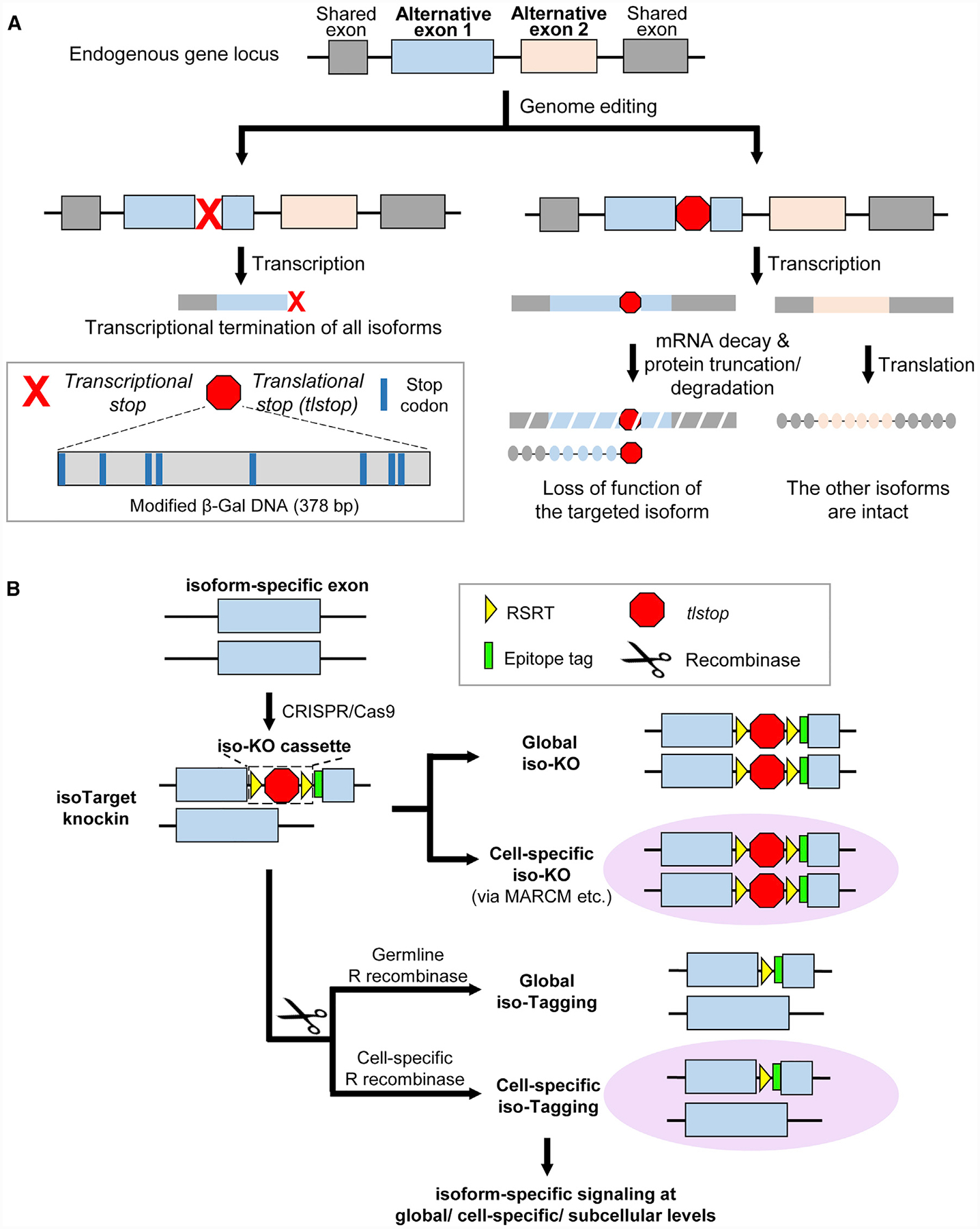
(A) Introducing translational stops into alternative exons allows isoform-specific manipulations of the gene. Inserting the commonly used transcriptional stop cassette into an alternative exon leads to transcriptional termination of all isoforms downstream of the targeted exon (left branch). To specifically manipulate one isoform (right branch), we engineered a translational stop (tlstop) cassette by introducing multiple stop codons into the DNA sequence encoding a non-catalytic region of β-Gal (bottom left). This tlstop cassette causes loss of function in targeted isoform by translational truncation or, possibly, mRNA decay.
(B) Combining tlstop with other genetic methods for multi-purpose studies of the targeted splicing isoform. The isoTarget cassette, which consists of an RSRT site, a translational stop (tlstop), a second RSRT site, and an epitope tag, is inserted into an isoform-specific exon by CRISPR/Cas9-mediated genome editing. The insertion is expected to generate a loss-of-function allele of the targeted isoform. Single cells that are homozygous for the targeted allele can be produced by genetic mosaic techniques, such as MARCM. The endogenous isoforms can be visualized in specific cells by selective expression of R recombinase to remove the RSRT-tlstop-RSRT cassette. Expression of R recombinase in female germline cells leads to tagging of the isoform in all cells that express that isoform in the progeny (“global iso-tagging”). Expression of R recombinase in specific cell types or single cells leads to tagging of the isoform in those cells. In the iso-tagged flies, upstream regulators and downstream effectors of specific isoforms can be identified through genetic, cell biological, and biochemical analyses.
See also Figures S1–S3 and S7.
