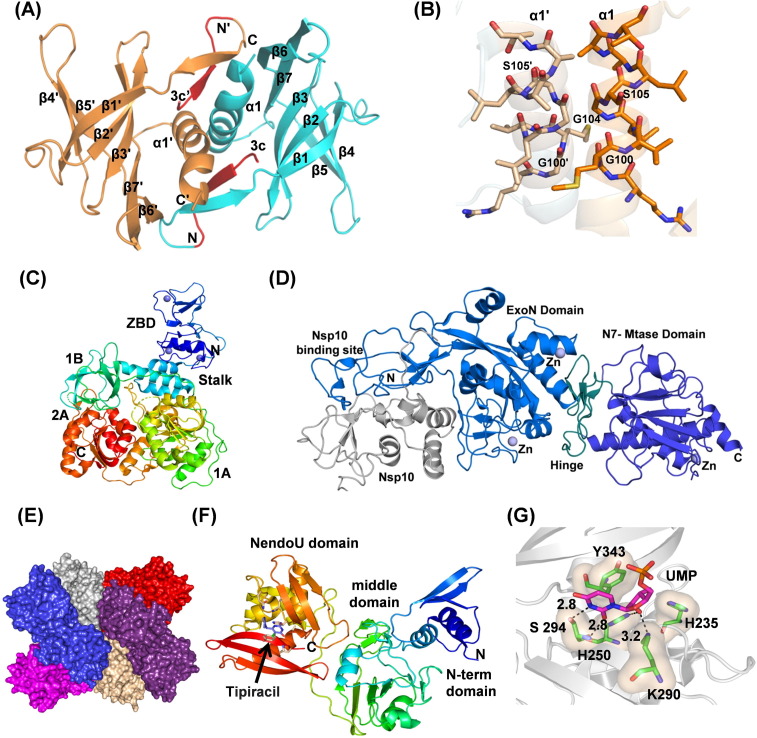
Fig. 5.
(A) Cartoon representation of Nsp9 (PDB ID: 6W9Q) showing dimer interface and peptide (shown in red) bound in the hydrophobic cavity. (B) Nsp9 showing protein–protein interacting helices with the GxxxG motif in stick representation. (C) The structure of Nsp13 (PDB ID: 6ZSL) showing zinc-binding domain (blue), stalk (blue-green), 1B (green), 1A (green-yellow) and 2A (red) domains. Three zinc atoms are shown as dark blue spheres. (D) Cartoon representation of SARS-CoV Nsp14-Nsp10 complex structure (PDB Id 5C8U) showing Nsp10 (gray), Nsp14 ExoN domain with nsp10-binding site (marine blue), hinge region (deep petal cyan), and the C-terminal N7-MTase domain (tv-blue). Zinc atoms are shown as spheres (slate). (E) Hexameric assembly of SARS-CoV-2 Nsp15 in surface representation. Each monomer is shown in different colors. (F) The structure of Nsp15 showing different domains (PDB ID: 6WXC). The active site residues are shown with the drug Tipiracil bound in the active site. (G) Binding of uridine monophosphate to the Nsp15 active site (PDB ID: 6WLC). Key conserved residues, H235, H250, K290, S294, and W343 are shown in a stick representation. S294 provides uridylate specificity. The hydroxyl group of S294 binds to the nitrogen atom of 5′ uracil while the main chain nitrogen atom interacts with the carbonyl oxygen.