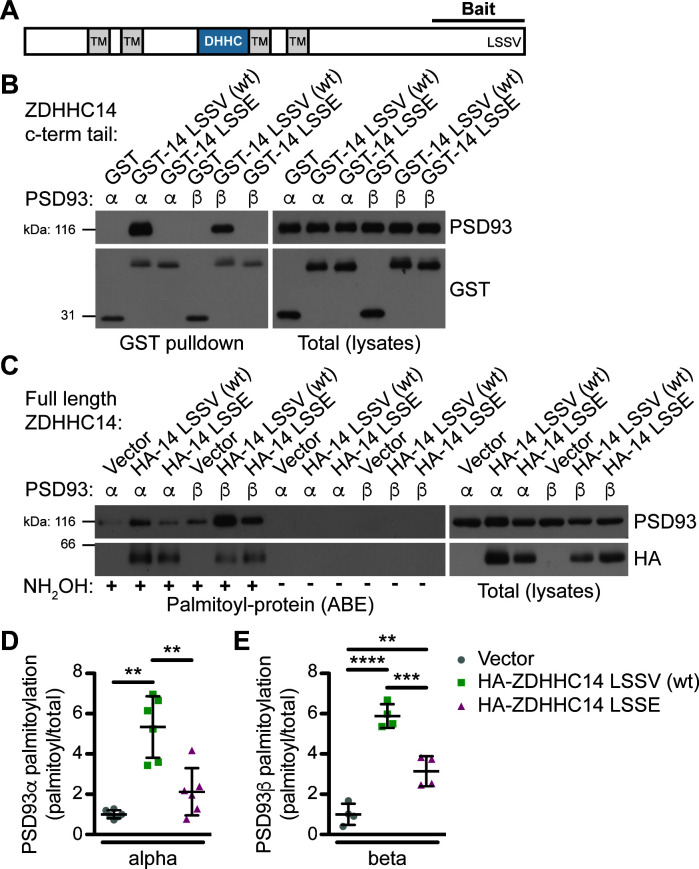
Figure 1. ZDHHC14 interacts with and palmitoylates both palmitoylated isoforms of PSD93 but more robustly palmitoylates PSD93β.
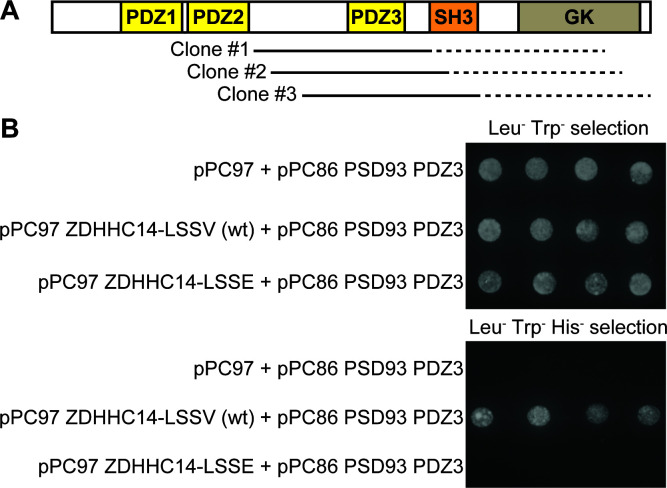
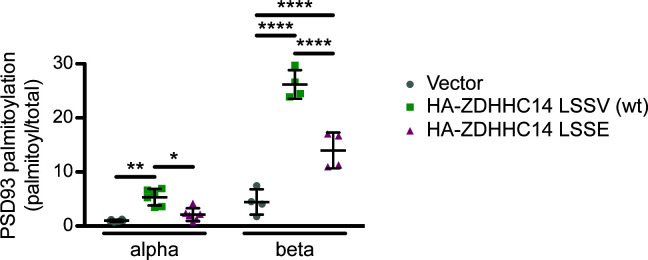
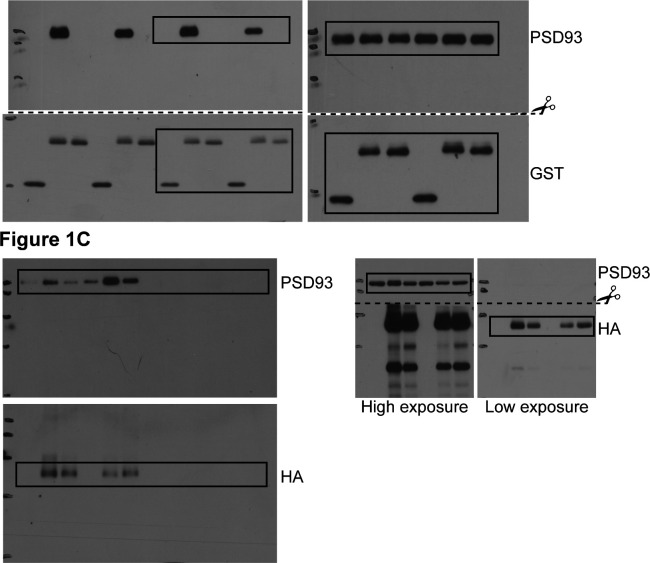
(A) Schematic of ZDHHC14 showing predicted transmembrane domains (TM, gray boxes), DHHC cysteine rich catalytic domain (blue box) and the C-terminal region used for yeast 2-hybrid screening (Bait), including the LSSV motif. (B) HEK293T cells were transfected with the indicated constructs and lysates subjected to GST-pulldown. Eluates from pulldowns were immunoblotted to detect GST (bottom, left) and PSD93 (top, left). Total expression levels of GST-tagged proteins (bottom, right) and PSD93 (top, right) in parent lysates were also determined. Images are representative of three independent experiments. (C) HEK293T cells were transfected with the indicated constructs and palmitoyl-proteins (isolated by ABE; left panels) and total protein levels (in parent lysates; right panels) were assessed by western blotting with the indicated antibodies. Parallel samples processed in the absence of the key ABE reagent hydroxylamine (NH2OH) confirm the specificity of the ABE assay. (D) Quantified PSD93α palmitoyl:total levels from C, normalized to the empty vector condition (Welch’s 1-way ANOVA p=0.0008, W(2,6.98) = 23.80, N = 6; Dunnett’s T3 multiple comparison post hoc test **p<0.01, 95% CI vector versus wtZDHHC14 [−6.91,–1.77], vector versus ZDHHC14 LSSE [−3.10, 0.86], and wtZDHHC14 versus ZDHHC14 LSSE [0.60, 5.83]). (E) Quantified PSD93β palmitoyl:total levels from C, normalized to the empty vector condition (1-way ANOVA p<0.0001, F(2,9)=60.69, N = 4; Bonferroni post hoc test **p<0.01, ***p<0.001, ****p<0.0001, 95% CI vector versus wtZDHHC14 [−6.18,–3.56], vector versus ZDHHC14 LSSE [−3.44,–0.84], and wtZDHHC14 versus ZDHHC14 LSSE [1.44, 4.04]). Uncropped western blot images are in Figure 1—figure supplement 4.