Abstract
The disease spectrum of coronavirus disease 2019 (COVID-19) ranges from no symptoms to multisystem failure and death. Characterization of virus-specific immune responses to severe acute respiratory coronavirus 2 (SARS–CoV-2) is key to understanding disease pathogenesis, but few studies have evaluated T cell immunity. In this issue of the JCI, Sattler and Angermair et al. sampled blood from subjects with COVID-19 and analyzed the activation and function of virus antigen–specific CD4+ T cells. T cells that failed to respond to peptides from the membrane, spike, or nucleocapsid proteins were more common in subjects who died. In those whose T cells had the capacity to respond, older patients with comorbidity had larger numbers of activated T cells compared with patients who had fewer risk factors, but these cells showed impaired IFN-γ production. This cross-sectional study relates activated T cell responses to patient risk factors and outcome. However, T cell response trajectory over the disease course remains an open question.
Introduction
The adaptive immune response is critical for virus clearance and eventual recovery from virus infection, but regulation of this response is required because inflammation can also contribute to immune-mediated tissue damage. T cell responses can help by promoting virus clearance and recovery or harm by causing immunopathology through direct tissue damage or excessive cytokine production. The role of T cells in the pathogenesis and clearance of severe acute respiratory coronavirus 2 (SARS–CoV-2) is of interest and importance, but cellular immune responses have been less well studied than plasma levels of antibodies, cytokines, and chemokines.
Previous studies of peripheral blood mononuclear cells (PBMCs) from acutely infected patients with coronavirus disease 2019 (COVID-19) have documented lymphopenia that primarily reflects decreased numbers of T lymphocytes in circulation (1–3). In general, T cells have multiple roles in the response to viral infection, including the cytotoxic effector roles of CD8+ T cells for clearance of virus-infected cells and production of soluble factors by a variety of CD4+ T cell subsets. CD4+ helper T cells function primarily by producing cytokines and chemokines that affect the differentiation, maturation function, and location of other cells both directly and indirectly. During COVID-19–induced lymphopenia, numbers of CD8+ cells in circulation are more severely decreased than CD4+ cells, resulting in elevated CD4/CD8 ratios (2, 4–6). Although decreased in number, both CD4+ and CD8+ T cells from patients with COVID-19 show evidence of recent proliferation and activation, indicating participation in an active cellular immune response to infection (1, 7, 8).
SARS–CoV-2 induces a wide spectrum of disease that spans asymptomatic infection and mild disease to fatal respiratory impairment with multisystem failure. Risk factors for more severe disease include older age, presence of comorbidities, and male sex (9–11). In general, those with asymptomatic infection develop less vigorous immune responses and clear the virus more slowly than those with mild disease (12). Severe disease is associated with more pronounced lymphopenia, greater plasma cytokine elevation, and more rapid virus clearance (1, 3, 5, 6, 9, 13, 14). However, early studies also suggested that CD4+ T cell production of interferon-gamma (IFN-γ), a T cell cytokine that has both antiviral and immune-stimulatory activities, and tumor necrosis factor-alpha (TNF-α) were impaired in patients with severe disease (14, 15).
These early studies suggest that, as with many other virus infections, COVID-19 is in large part a manifestation of the immune response to SARS–CoV-2 and that specifics of the immune response determine outcome. Therefore, understanding the role of cellular immunity in outcome from infection is of critical importance both for identifying therapies and developing a vaccine. Importantly, understanding the relationship between cellular immunity and infection outcome requires detailed study of immune effectors as well as characterization of viral antigen–specific responses.
Viruses produce many proteins for replication and regulation of cell processes, as well as for assembly of new virus particles. The immune system can recognize these viral proteins as foreign, but innate and adaptive immune responses to each protein will vary in their impact on disease pathogenesis, virus clearance, and protection from reinfection. Therefore, analysis of antigen-specific responses is essential to understand the determinants of outcome, as well as the requirements for inducing vaccine immunity.
The SARS–CoV-2 virus particle is comprised of four proteins: envelope (E), spike (S), and membrane (M) are transmembrane envelope proteins on the virion surface, and nucleoprotein (N) encapsidates the viral genomic RNA inside the particle. In addition, the virus encodes more than 20 nonstructural replicase and regulatory proteins (16, 17). The adaptive immune response can target all of these proteins. T cells recognize peptides derived from proteins expressed by virus-infected cells and viral proteins taken up and processed by antigen presenting cells. Selected peptides are presented in association with major histocompatibility complex I or II, and bioinformatic analyses have identified likely T cell epitopes in multiple SARS–CoV-2 proteins, particularly S, M, and N (18).
Are T cells helping?
CD4+ Th cells are functionally diverse. Several subsets have been defined by transcription factor and cytokine and chemokine expression, but there is substantial plasticity, and overlapping phenotypes are common (19, 20). Th1 cells expressing IFN-γ, TNF-α, and IL-2 are classically considered most important for the response to intracellular pathogens, including viruses. However, virus infections induce many other types of CD4+ T cells with relevant functions (21). For instance, Th2 cells expressing IL-4, IL-5, and IL-13 provide help for B cell proliferation. Th17 cells expressing IL-17 and IL-22 can act pathogenic or regulatory depending in part on whether they also produce IFN-γ or IL-10. T follicular helper (Tfh) cells express IL-21 to promote germinal center formation for maturation of antibody-secreting cells. Regulatory T cells express IL-10. All of these lineages can be identified in peripheral blood in varying proportions during the response to infection, although the primary sites of function are in lymphoid tissues and at sites of virus replication.
In this issue of the JCI, Sattler and Angermair et al. (22) used flow cytometry analysis of PBMCs to examine the CD4+ T cell responses to stimulation with overlapping peptides from SARS–CoV-2 S, M, and N proteins for 39 hospitalized patients stratified by intensive care unit status and seven nonhospitalized patients recovered from mild disease. Virus-specific T cells, as identified by surface expression of activation markers after stimulation in culture with viral peptides, were present in most acutely ill hospitalized patients and comparable numbers of CD4+ T cells responded to peptides from each of the proteins (22).
Patients who failed to mount a CD4+ T cell response (nonresponders) were primarily in the group sampled early in their disease course. While these subjects showed similar T cell numbers with those who successfully mounted a T cell response (responders), S, M, and N failed to stimulate the T cells themselves (22). An early deficiency in CD4+ T cell response is consistent with the previously reported two-week time frame of symptom onset, and for the development of the CD4+ T cell response to S (4). The response to S is of particular interest because the spike protein is the viral surface glycoprotein that contains the binding site for the ACE-2 cellular receptor and is the target for neutralizing antibodies and the focus of most vaccine development efforts. Patients with no evidence of a CD4+ T cell response to S peptides were also negative for IgG antibody to S in serum, suggesting a failure to mount either an effective B cell or T cell adaptive immune response to this viral protein. These nonresponding patients were also most likely to die from COVID-19, further indicating the importance of prompt virus-specific immune responses for recovery (22).
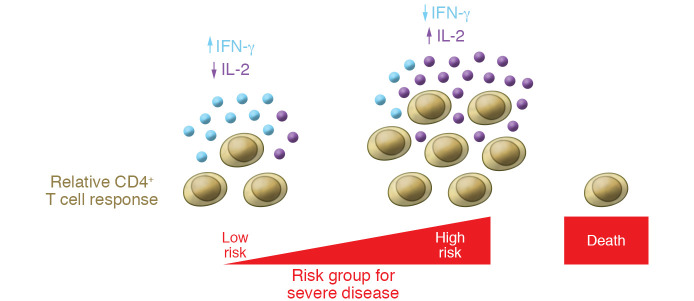
Interestingly, among responding patients, higher CD4+ T cell responses to all three proteins were associated with older age, higher comorbidity indices, and more severe disease with the most substantial differences in responses to M. To determine the functional capacity of the CD4+ T cells responding to each of the viral proteins, Sattler, Angermair, and colleagues measured IL-2, TNF-α, and IFN-γ production. N-specific cells were least likely, and M-specific T cells were most likely, to produce IFN-γ alone or in combination with TNF-α. In addition to antigen-specific differences in function, differences among patient groups were also evident. In older patients with higher comorbidity indices, even though there were larger numbers of cells, activated cells were less likely to secrete IFN-γ and more likely to produce IL-2 (Figure 1). However, differences between patients with different levels of disease severity were not evident (22). This relationship contrasts with earlier studies of CD4+ T cell function that identified lower percentages of cells producing IFN-γ in severely ill patients compared with those having mild disease, though viral protein specificity was not determined (14).
Figure 1. Model for T cell response in COVID-19 relative to patient risk.
Patients at risk for severe COVID-19 disease (advanced age and higher comorbidity) show higher T cell responses than those at low risk. However, the activated T cells from patients at risk have impaired IFN-γ and enhanced IL-2 production.
Study limitations and future directions
This cross-sectional study provides important data on CD4+ T cell responses to three different immunogenic SARS–CoV-2 structural proteins and the relationship of these responses to patient risk factors and outcome (22). However, there is no information on the trajectory of these responses over the course of disease progression or recovery. Previous studies show that T cells can recover rapidly as virus is cleared (6), and there were substantial differences in time after infection for samples taken from the recovered and hospitalized patients.
In addition, only Th1 cells were evaluated. A decreased proportion of IFN-γ–secreting Th1 cells despite evidence of CD4+ T cell activation may suggest that the activated CD4+ T cells belong to a different subset of helper T cells than Th1 (22). Other studies of hospitalized patients with COVID-19 indicate that both Th17 and regulatory T cells are present in the activated CD4+ T cell populations (8) and that PBMCs produce cytokines characteristic of Th2 and Th17 as well as Th1 cells (4). Furthermore, during convalescence when lymphocyte counts rebound there is an increase in memory and S-reactive CD4+ T cells (23), many of which have been identified as Tfh cells (24).
In summary, this study provides valuable information on viral antigen–specific CD4+ T cell responses and serves as a basis for further studies of CD4+ T cell function in determining outcome of SARS–CoV-2 infection. Because T cell as well as antibody responses increase over time (4), longitudinal analysis of patients with both mild and severe disease would be valuable to determine how the function of viral protein–specific T cells evolves with time after infection in patients with different manifestations.
Acknowledgments
DEG is supported by research grants from the National Institutes of Health (R01 AI131228, R01 AI153140, R56 AI137264), Gilead Pharmaceuticals, and the Johns Hopkins COVID-19 Research Program.
Version 1. 09/25/2020
In-Press Preview
Version 2. 11/03/2020
Electronic publication
Version 3. 12/01/2020
Print issue publication
Footnotes
Conflict of interest: DEG receives research funding from Gilead Pharmaceuticals and is a member of the Vaccines Research and Development Advisory Board for GlaxoSmithKline and the Zika Vaccine Data Monitoring Committee for Takeda Pharmaceuticals.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(12):6222–6224. https://doi.org/10.1172/JCI142081.
See the related article at SARS–CoV-2–specific T cell responses and correlations with COVID-19 patient predisposition.
References
- 1.Song JW, et al. Immunological and inflammatory profiles in mild and severe cases of COVID-19. Nat Commun. 2020;11(1):3410. doi: 10.1038/s41467-020-17240-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schultheiß C, et al. Next-generation sequencing of T and B cell receptor repertoires from COVID-19 patients showed signatures associated with severity of disease. Immunity. 2020;53(2):442–455.e4. doi: 10.1016/j.immuni.2020.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qin C, et al. Dysregulation of immune response in patients with coronavirus 2019 (COVID-19) in Wuhan, China. Clin Infect Dis. 2020;71(15):762–768. doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weiskopf D, et al. Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci Immunol. 2020;5(48):eabd2071. doi: 10.1126/sciimmunol.abd2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang F, et al. Characteristics of peripheral lymphocyte subset alteration in COVID-19 pneumonia. J Infect Dis. 2020;221(11):1762–1769. doi: 10.1093/infdis/jiaa150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang M, et al. T-cell subset counts in peripheral blood can be used as discriminatory biomarkers for diagnosis and severity prediction of coronavirus disease 2019. J Infect Dis. 2020;222(2):198–202. doi: 10.1093/infdis/jiaa252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Laing AG, et al. A dynamic COVID-19 immune signature includes associations with poor prognosis. Nat Med. 2020;26(10):1623–1635. doi: 10.1038/s41591-020-1038-6. [DOI] [PubMed] [Google Scholar]
- 8.De Biasi S, et al. Marked T cell activation, senescence, exhaustion and skewing towards TH17 in patients with COVID-19 pneumonia. Nat Commun. 2020;11(1):3434. doi: 10.1038/s41467-020-17292-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang C, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang D, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323(11):1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou F, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Long QX, et al. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat Med. 2020;26(8):1200–1204. doi: 10.1038/s41591-020-0965-6. [DOI] [PubMed] [Google Scholar]
- 13.Xu B, et al. Suppressed T cell-mediated immunity in patients with COVID-19: A clinical retrospective study in Wuhan, China. J Infect. 2020;81(1):e51–e60. doi: 10.1016/j.jinf.2020.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen G, et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J Clin Invest. 2020;130(5):2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng HY, et al. Elevated exhaustion levels and reduced functional diversity of T cells in peripheral blood may predict severe progression in COVID-19 patients. Cell Mol Immunol. 2020;17(5):541–543. doi: 10.1038/s41423-020-0401-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu F, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou P, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grifoni A, Sidney J, Zhang Y, Scheuermann RH, Peters B, Sette A. A sequence homology and bioinformatic approach can predict candidate targets for immune responses to SARS-CoV-2. Cell Host Microbe. 2020;27(4):671–680.e2. doi: 10.1016/j.chom.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sallusto F. Heterogeneity of human CD4(+) T cells against microbes. Annu Rev Immunol. 2016;34:317–334. doi: 10.1146/annurev-immunol-032414-112056. [DOI] [PubMed] [Google Scholar]
- 20.Nakayamada S, Takahashi H, Kanno Y, O’Shea JJ. Helper T cell diversity and plasticity. Curr Opin Immunol. 2012;24(3):297–302. doi: 10.1016/j.coi.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beňová K, Hancková M, Koči K, Kúdelová M, Betáková T. T cells and their function in the immune response to viruses. Acta Virol. 2020;64(2):131–143. doi: 10.4149/av_2020_203. [DOI] [PubMed] [Google Scholar]
- 22.Sattler A, et al. SARS–CoV-2–specific T cell responses and correlations with COVID-19 patient predisposition. J Clin Invest. 2020;130(12):6477–6489. doi: 10.1172/JCI140965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grifoni A, et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181(7):1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Juno JA, et al. Humoral circulating follicular helper T cell responses in recovered patients with COVID-19. Nat Med. 2020;26(9):1428–1434. doi: 10.1038/s41591-020-0995-0. [DOI] [PubMed] [Google Scholar]