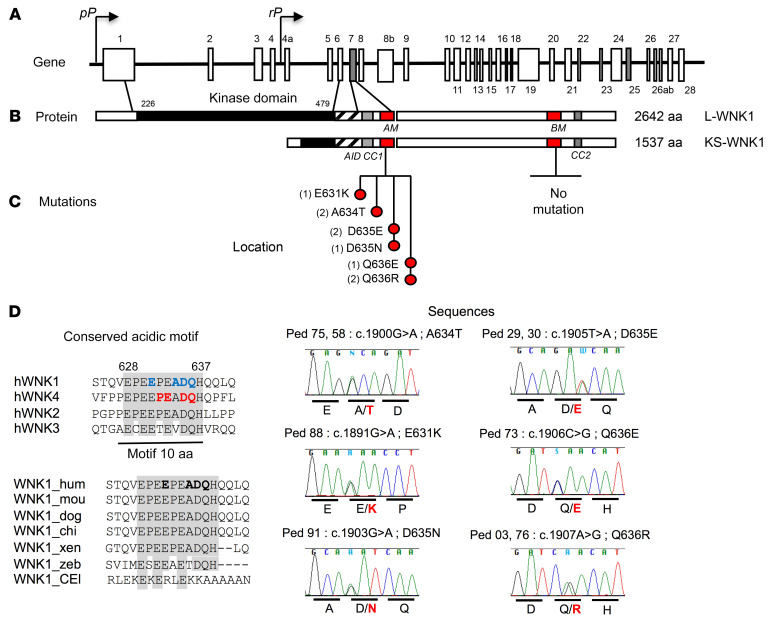
Figure 2. Acidic motif WNK1 mutations.
(A) Schematic representation of the WNK1 gene. The coordinates of the different exons are indicated above or below the structure with ex7 and ex25 (gray), which code for the conserved acidic and basic motifs. The proximal promoter (pP) drives the expression of the long ubiquitous kinase active isoform (L-WNK1), whereas the renal promoter (rP) drives the expression of the kinase defective kidney-specific isoform (KS-WNK1). (B) Schematic linear structure of LWNK1 and KS-WNK1. The kinase domain is represented in full black, the autoinhibitory domain (AID) is striped, coiled-coil domain 1 (CC1) and CC2 are represented in gray, and the conserved acidic motif (AM) and basic motif (BM) are represented in red. (C) Location and sequences of the mutated residues clustering in the acidic motif. The brackets indicate the number of unrelated affected subjects for each mutation. On the lower right are shown the Sanger sequencing electrophoregrams showing the various missense WNK1 mutations. (D) Conservation of the acidic motif showing residues mutated in FHHt among human WNK family members. The previously described WNK4 mutations are indicated in red; those at WNK1 and identified in this study in blue. All are located at completely conserved residues. The bottom part shows the conservation of the WNK1 acidic motif across species. The mutated residues are indicated in bold.