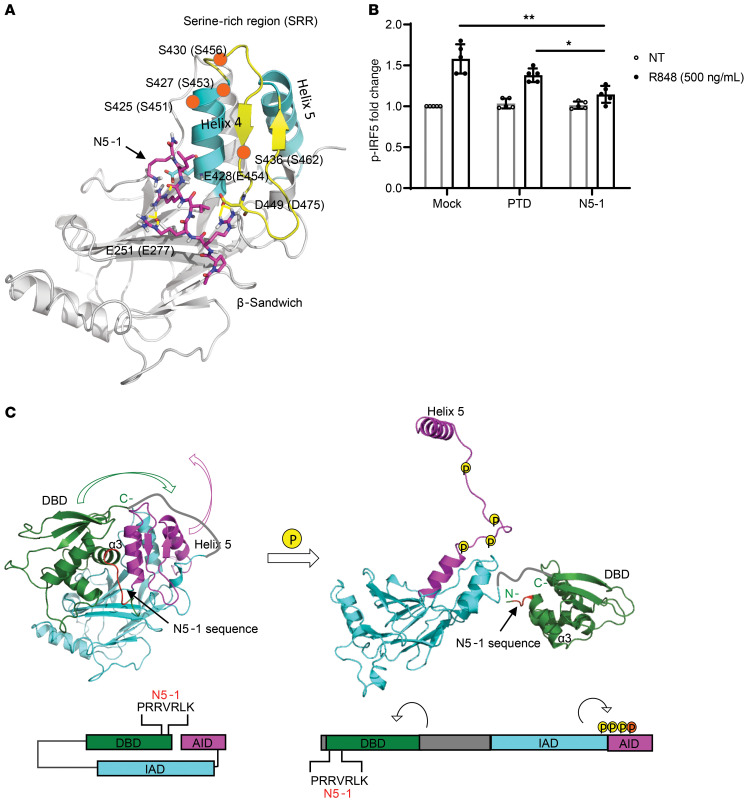
Figure 3. N5-1 is predicted to bind to the C-terminal IAD of an inactive IRF5 monomer and inhibit phosphorylation of Ser462.
(A) Schematic diagram of N5-1 (pink) binding to the C-terminal IAD of IRF5 from peptide docking using the Schrodinger suite (see Supplemental Methods). N5-1 stabilizes the nonphosphorylated, inactive IRF5 monomer. Serine phosphorylation sites are shown by orange circles. (B) PBMCs were preincubated with 10 μM inhibitor for 1 hour and stimulated with R848. p-IRF5 phosphorylation at Ser462 was detected by flow cytometry following gating on CD14+ monocytes. The fold change in p-IRF5 relative to unstimulated mock samples is shown. n = 5 independent samples from healthy donors. Data represent the mean ± SD. *P ≤ 0.05 and **P ≤ 0.01, by 1-way ANOVA. (C) On the basis of the binding of N5-1 to full-length inactive IRF5, we propose that the DBD masks the IAD of IRF5 and that the AID masks the C-terminal phosphorylation sites, thus stabilizing a closed, unphosphorylated conformation of the IRF5 monomer (left panel). In this conformation, the DBD α3 helix, which contains all the conserved residues and is responsible for protein-DNA contacts, is shielded. Upon phosphorylation, the AID unfolds, which unmasks the C-terminal phosphorylation sites and frees helix 5 for dimerization (right panel). The DBD will also be released from this folded, inactive position and exposed to DNA for binding. The colors correspond to the specified regions of IRF5 in the crystal structure (above) and the stick model (below). The DBD is indicated in green, the IAD in blue, and the AID in purple. The N5-1 sequence is shown in red in both models.