ABSTRACT
Chromosomes are dynamic entities, whose organization and structure depend on the concerted activity of DNA-binding proteins and DNA-processing enzymes. In bacteria, chromosome replication, segregation, compaction and transcription are all occurring simultaneously, and to ensure that these processes are appropriately coordinated, all bacteria employ a mix of well-conserved and species-specific proteins. Unusually, Streptomyces bacteria have large, linear chromosomes and life cycle stages that include multigenomic filamentous hyphae and unigenomic spores. Moreover, their prolific secondary metabolism yields a wealth of bioactive natural products. These different life cycle stages are associated with profound changes in nucleoid structure and chromosome compaction, and require distinct repertoires of architectural—and regulatory—proteins. To date, chromosome organization is best understood during Streptomyces sporulation, when chromosome segregation and condensation are most evident, and these processes are coordinated with synchronous rounds of cell division. Advances are, however, now being made in understanding how chromosome organization is achieved in multigenomic hyphal compartments, in defining the functional and regulatory interplay between different architectural elements, and in appreciating the transcriptional control exerted by these ‘structural’ proteins.
Keywords: Streptomyces, nucleoid-associated proteins, topoisomerase, chromosome domain, chromosome organization, chromosome topology, sporulation, regulation of gene expression, condensin
Streptomyces employ distinct repertoires of proteins to organize their chromosomes throughout their complex life cycle.
INTRODUCTION
Bacterial chromosomes are dynamic macromolecules that are subject to multilevel organization; however, the basic chromosomal unit can vary between bacteria. Most bacteria possess a single, circular chromosome. There are, however, bacteria that have linear chromosomes, as well as ones that have multiple chromosomes per cell. In all cases, these chromosomes are compacted into ‘nucleoid’ structures within the cytoplasm (Murphy and Zimmerman 1995; Feijoo-Siota et al. 2017; Dame, Rashid and Grainger 2019). While extensive compaction is essential in order to accommodate the chromosome within a cell, chromosome organization must be flexible enough to allow for simultaneous DNA replication, chromosome segregation and transcription.
Compact, dynamic nucleoids are maintained by a set of DNA-organizing proteins including those that are highly conserved (e.g. topoisomerases) (Forterre et al. 2007), and others that are unique to particular bacterial groups (Dame, Rashid and Grainger 2019). In the case of Streptomyces, these soil-dwelling, antibiotic-producing bacteria have linear chromosomes (Kirby 2011), and an unusual multicellular life cycle with stages that include multichromosomal hyphal filaments (Flärdh et al. 2012) and single genome spore compartments (Elliot and Flärdh 2012). Consequently, these organisms face unique challenges in terms of chromosome replication, segregation and condensation, as well as the need to coordinate these processes with the different stages of their life cycle (Flärdh and Buttner 2009; Jones and Elliot 2018). Here, we describe the chromosome properties and processes that are conserved throughout bacteria, and discuss how streptomycetes achieve dynamic chromosome organization and circumvent the challenges associated with their large (8–13 Mbp), linear chromosome and their complex life cycle. We further discuss the role of chromosome topology in controlling gene expression, stress adaptation and antibiotic production in these remarkable bacteria.
CHROMOSOME ORGANIZATION—GENERAL CHARACTERISTICS AND WELL-STUDIED EXAMPLES
Bacterial chromosomes tend to adopt a loosely twisted conformation that is dictated by macromolecular crowding (de Vries 2010), and initially, these molecules were not expected to be organized in a defined manner. Chromosome orientation was first probed using fluorescent labels that allowed for the localization of the origin (oriC) and terminus (ter); the positioning of each turned out to be both dynamic and predictable over the course of a cell cycle (Webb et al. 1997; Niki and Hiraga 1998; Espéli and Boccard 2006; Espeli, Mercier and Boccard 2008). Intriguingly, the specific orientations of the oriC and ter loci were found to be organism specific (Wang, Montero Llopis and Rudner 2013; Badrinarayanan, Le and Laub 2015). Many bacteria position their oriC and ter regions at opposite poles of the newborn cell (e.g. Caulobacter cresentus, Vibrio cholerae), while others have evolved a subpolar oriC location (e.g. Myxococcus xanthus, Mycobacterium smegmatis) (Ebersbach et al. 2008; Harms et al. 2013; David et al. 2014; Hołówka et al. 2018). Growth rate and life cycle stage can also influence oriC positioning. For example, in slow-growing Escherichia coli, the oriC is located at mid-cell, while in fast-growing cultures, it is located at the cell pole (Niki, Yamaichi and Hiraga 2000; Kleckner et al. 2014; Badrinarayanan, Le and Laub 2015). In Bacillus subtilis, life cycle stage impacts the configuration of the chromosome, with oriC shifting from a central position during vegetative growth, to a polar location at the onset of sporulation (Errington 2001; Wang, Montero Llopis and Rudner 2013; Badrinarayanan, Le and Laub 2015).
In bacteria with a polarly localized oriC, the onset of DNA replication is followed by the translocation of one copy of the oriC to the opposite pole. In contrast, in cells with subpolar or central oriC orientations, both oriC copies move toward opposite poles (Ben-Yehuda, Rudner and Losick 2003; Ebersbach et al. 2008; Wang, Montero Llopis and Rudner 2014). In all cases, however, the newly replicated oriC(s) reach their final destination (pole, subpole or mid-cell) before replication is complete. In most bacterial species (except for the γ-proteobacteria), oriC positioning is dictated by a chromosome segregation system comprising the ParA and ParB proteins, and the parS DNA sequences (Lim et al. 2014; Badrinarayanan, Le and Laub 2015; Kawalek et al. 2020). ParA is an ATPase that dimerizes and binds DNA non-specifically when in an ATP-bound configuration, while ParB is a DNA-binding protein that specifically recognizes and binds multiple parS sequences that flank the oriC; these parS sequences are distributed over an area encompassing anywhere from 3% (in Helicobacter pylori) to 20% (in Bacillus subtilis) of the total chromosome (Lin and Grossman 1998; Livny, Yamaichi and Waldor 2007). Upon binding these parS sequences, ParB is proposed to spread non-specifically along the DNA and bridge distant chromosomal regions in a CTP-dependent manner (Osorio-Valeriano et al. 2019; Soh et al. 2019; Jalal, Tran and Le 2020), ultimately forming a large nucleoprotein complex known as the segrosome (Graham et al. 2014; Song et al. 2017). Segrosomes interact with nucleoid-bound ParA, and this interaction stimulates ParA ATPase activity, triggering protein release from DNA. Thus, the presence of segrosomes results in the formation of a ParA-depleted zone, and this in turn drives ParB complexed-DNA toward higher concentrations of nearby nucleoid bound ParA (Vecchiarelli, Neuman and Mizuuchi 2014; Surovtsev, Campos and Jacobs-Wagner 2016). Notably, segregation proteins not only control the subcellular position of the oriC region but also organize this region in a way that facilitates its segregation.
It has been more challenging to follow the specific localization and organization of the chromosome arms (between oriC and ter); however, recent innovations in fluorescent microscopy and chromosome conformation capture technologies are revealing reproducible architectures for these regions as well (Niki, Yamaichi and Hiraga 2000; Valens et al. 2004; Viollier et al. 2004; Lioy et al. 2018). In E. coli, the chromosome arms, as well as the oriC and ter regions, are organized into well-compacted macrodomains that are separated by unstructured DNA regions (Valens et al. 2004). The principles governing macrodomain organization in E. coli are poorly understood, with the exception of the oriC and ter macrodomains, whose spatial architecture are dictated by the MaoP and MatP proteins, respectively (Mercier et al. 2008; Valens, Thiel and Boccard 2016). MaoP and MatP bind and bridge their cognate maoS/matS DNA sequences, which are distributed along ∼800 kb DNA fragments. MaoP and MatP homologs are, however, restricted to the enteric bacteria, and consequently, it is not yet clear whether such organization is limited to E. coli and its close relatives, or whether other bacteria have evolved analogous systems for organizing their origin and terminus regions.
In bacterial species with polar oriC (and ter) configurations, the left and right chromosomal arms remain in close proximity throughout the cell cycle. This is thought to result from long-distance arm cohesion driven by DNA condensins (Wang et al. 2017) that function using a loop extrusion mechanism (Marko et al. 2019). In most bacteria, condensin complexes are composed of SMC (structural maintenance of chromosomes), a large coiled-coil protein dimer and two partner proteins, ScpA and ScpB (segregation and condensation proteins). These SMC/ScpAB complexes are recruited to DNA through interaction with the segregation protein ParB (Gruber and Errington 2009; Sullivan, Marquis and Rudner 2009; Minnen et al. 2011). Thus, the ParB nucleoprotein complex is essential for not only organizing and segregating oriC regions but also facilitating SMC loading, which in turn impacts global chromosome compaction. Interestingly, in E. coli (which lacks ParAB proteins), recruitment of the SMC homolog MukB is controlled by the ter-associated MatP protein (Nolivos et al. 2016; Mäkelä and Sherratt 2020). Moreover, MukB also interacts with topoisomerase IV during oriC segregation (Hayama and Marians 2010; Nolivos et al. 2016).
Locally, bacterial chromosomes are organized into specific domains that differ in both size and spatial architecture. In Caulobacter crescentus, chromosome conformation capture experiments have revealed the chromosome is divided into >20 ‘chromosomal interaction domains’ (CIDs), each averaging ∼160 kb in size (Le et al. 2013). The boundaries of particular domains often coincide with highly expressed genes, suggesting that transcription may make a major contribution to the integrity of each CID and their subsequent spatial organization into higher order structures similar to E. coli macrodomains (Badrinarayanan, Le and Laub 2015; Shen and Landick 2019). Within any given CID, the DNA is organized into plectonemically supercoiled loops called ‘topological domains’ or ‘supercoiling domains’, having an estimated average size of ∼10 kb (Postow et al. 2004). Like the larger domains (macrodomains and CIDs), the intrinsic architecture of topological domains is maintained by nucleoid associated proteins (NAPs) and topoisomerases, as well as by the transcriptional activity of genes located within the particular domains. NAPs are a heterogeneous group of small, basic, often dimeric proteins that bind DNA with varying degrees of sequence specificity (Hardy and Cozzarelli 2005). Like eukaryotic histones, NAPs are responsible for DNA bending (HU, IHF or Fis homologs), wrapping (Lrp and Dps homologs) or local bridging of DNA fragments (H-NS-like proteins), and all are proposed to stabilize topological domains or modulate their architecture (Badrinarayanan, Le and Laub 2015; Dame, Rashid and Grainger 2019). NAPs can also profoundly affect gene transcription (Dorman 2013; Dorman et al. 2020). Notably, the repertoire of NAPs employed by different bacterial species can be highly variable (Table 1).
Table 1.
Conservation of select proteins having roles in chromosome dynamics in phylogenetically diverse bacteria.
| E. coli | B. subtilis | C. crescentus | M. tuberculosis | S. coelicolor | |
|---|---|---|---|---|---|
| Genome content | AT-rich | AT-rich | GC-rich | GC-rich | GC-rich |
| Gram +/− | Gram-negative | Gram-positive | Gram-negative | Gram-positive | Gram-positive |
| Protein: | |||||
| HU | HU (α and β subunits) | Hbsu | HCc | HupB (Rv2986) | HupA, HupS |
| DpsA | Dps | Dps | Dps | – | DpsA, DpsB, DpsC |
| Fis | Fis | – | – | – | – |
| IHF homologs and functional equivalents | IHF (α and β subunits) | IHF | mIHF (functional equivalent) | sIHF (functional equivalent) | |
| H-NS and H-NS-like proteins | H-NS, StpA (H-NS paralog) | Rok (functional equivalent) | GapRa (functional equivalent) | Lsr2 (functional equivalent) | Lsr2 (functional equivalent) |
| ParA/ParB | – | Soj/Spo0J | ParAB | ParAB | ParAB |
| Noc | – | Noc | – | – | – |
| Condensin | MukB | SMC | SMC | SMC | SMC |
| MaoP | MaoP | – | – | – | – |
| MatP | MatP | – | – | – | – |
| TopA (TopoI) | TopA | TopA | TopA | TopA | TopA |
| TopB (TopoIII) | TopB | TopB | – | – | – |
| GyrAB (gyrase) | GyrA and GyrB | GyrBA | GyrA and GyrB | GyrBA | GyrBA |
| ParCE (TopoIV) | ParC and ParE | ParC and ParE | ParC and ParE | – | ParC and ParE |
| Ssb | Ssb | SsbA and SsbB | – | Ssb | SsbA and SsbB |
GapR is found in the α-proteobacteria and, like H-NS, has an affinity for AT-rich DNA (Ricci et al. 2016).
Gray shading: essential genes. For species with SsbA and SsbB, typically only SsbA is essential.
Topological domain compaction is also controlled by topoisomerases; these enzymes are responsible for adding or removing DNA supercoils through the breaking and rejoining of phosphodiester bonds. Topoisomerases are critical for removing the supercoils that result from DNA replication and transcription, and further facilitate DNA strand separation during recombination (Brochu, Breton and Drolet 2020). Two topoisomerases are essential for viability and are present in all bacteria: TopA, which removes negative supercoils from DNA, and gyrase, which introduces negative supercoils. The opposing activities of TopA and gyrase are crucial for maintaining the appropriate balance between supercoiling-driven DNA compaction, and DNA accessibility—a phenomenon known as topological homeostasis (Ahmed et al. 2015; Ferrandiz et al. 2016; Szafran et al. 2016). Interestingly, some nucleoid-associated proteins (e.g. the mycobacterial HU homolog HupB) can influence topoisomerase activity through their changing of DNA spatial structure, or through direct protein–protein interactions (Ghosh, Mallick and Nagaraja 2014). Nucleoid-associated proteins can also limit the diffusion or spread of DNA supercoils by establishing local barriers, ultimately leading to the generation of distinct supercoiling gradients along the bacterial chromosome (Lal et al. 2016; Ferrándiz et al. 2018). Overall, the organization of topological domains is dynamic and relies on the combined activities of DNA-binding proteins and DNA-processing enzymes.
Streptomyces spp. employ the same chromosome organization systems as many other bacteria, including the ParABS system for chromosome segregation, alongside dedicated NAPs, SMC-type condensins and topoisomerases. As in other systems, the activity of the various chromosome-organizing proteins in Streptomyces depends on their life cycle stage. In unicellular and unigenomic bacteria, chromosome organization is tightly coordinated with cell cycle and cell shape. Given the unusual nature of the filamentous and sporulating streptomycetes, the chromosome-organizing machineries in these bacteria must deal with multiple chromosomes in the vegetative and aerial hyphal filaments, as well as the synchronous segregation and compaction of these chromosomes during sporulation, to ensure that each spore inherits a single chromosome.
STREPTOMYCES LIFE CYCLE
The Streptomyces life cycle begins with a unigenomic spore (Fig. 1A). Spore germination initiates with the emergence of one or two germ tubes at sites marked by the polarity determinant DivIVA (Flärdh 2003a) (Fig. 1B). Unlike most other bacteria, Streptomyces do not undergo binary fission. Instead they grow filamentously via polar tip extension (Gray, Gooday and Prosser 1990), and forgo cell division except during sporulation (McCormick 2009). Extension of these initial germ tubes yields long hyphal filaments, from which periodic hyphal branches emerge. This branching, hyphal growth strategy ultimately leads to the formation of a network of filamentous vegetative cells, each of which contains multiple, largely uncondensed chromosomes (Kwak and Kendrick 1996) (Fig. 1C).
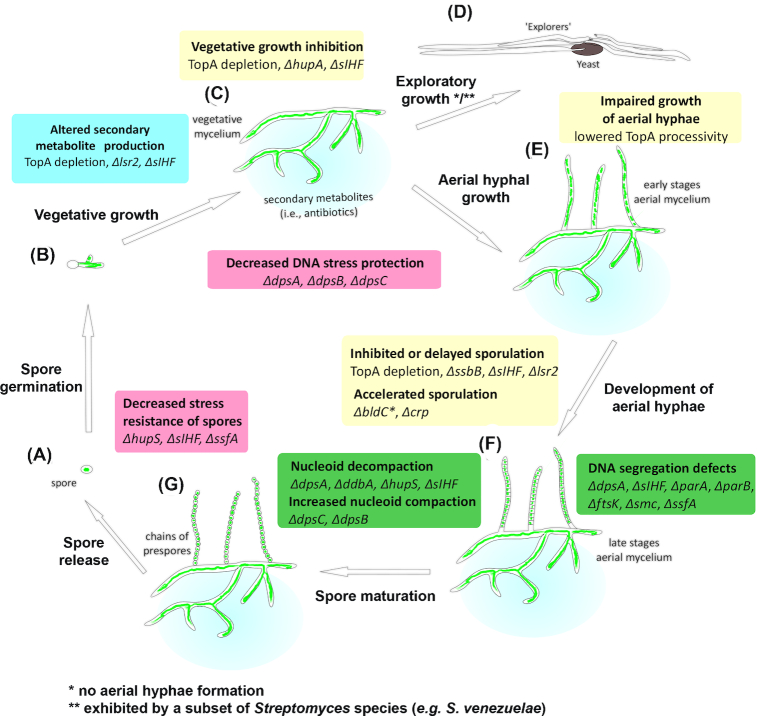
Figure 1.
The role of DNA-processing proteins during the Streptomyces life cycle. The Streptomyces life cycle begins with single spore (A), which germinates (B) and grows via tip extension and branching to form a network of vegetative hyphae (C). Nutrient depletion or environmental stress induces exploratory growth (D) or sporulation initiated by aerial hyphal growth (E). The aerial hyphae mature into chains of pre-spores (F) and subsequently spores (G) that are disseminated. The phenotypic effects of mutations in genes encoding chromosome-organizing proteins are indicated. Pink boxes: altered stress resistance. Blue box: modified secondary metabolism. Yellow boxes: modified growth. Green boxes: altered chromosome topology.
Hyphal tip growth is governed by DivIVA, which directs cell wall biosynthesis and is essential for Streptomyces viability (Flärdh 2003a). Beyond its role in coordinating peptidoglycan biosynthesis, DivIVA acts in concert with a number of other proteins at the hyphal tip. These tip-associated proteins are collectively referred to as the ‘polarisome’, and they include the intermediate filament-like protein FilP, which interacts directly with DivIVA (Fuchino et al. 2013; Fröjd and Flärdh 2019); and the coiled-coil protein Scy, which interacts with both DivIVA and FilP, as well as with ParA—the chromosome segregation-associated protein (Ditkowski et al. 2013; Holmes et al. 2013). The interaction between ParA and Scy couples hyphal tip growth with chromosome segregation in the growing vegetative hyphae (Kois-Ostrowska et al. 2016).
Branch formation is initiated when the polarisome splits, and a new polarisome complex assembles along the lateral cell wall, driving new tip growth (Hempel et al. 2008; Flärdh et al. 2012; Richards et al. 2012). Importantly, while the activity of DivIVA and its associated proteins are required for branch formation, continued branch growth requires the successful capture/translocation of a chromosome into the nascent branch; if a branch fails to acquire a chromosome, its elongation ceases (Kois-Ostrowska et al. 2016).
When nutrients are abundant, vegetative growth continues. However, when resources become scarce, or when stressful conditions are encountered, Streptomyces colonies embark on one of two developmental trajectories: (i) reproductive growth, involving the raising of aerial hyphae and formation of spore chains (Elliot and Flärdh 2012) or (ii) exploratory growth, involving rapid colony expansion and an ability to colonize new areas (Jones et al. 2017) (Fig. 1D and E).
Aerial hyphal growth, like that of the vegetative hyphae, is driven by DivIVA and its associated polarisome constituents, although branching is not observed in the aerial hyphae (Fig. 1E). The growth of aerial hyphae is closely coupled to chromosome replication and segregation (discussed below), and the cessation of growth is correlated with the onset of sporulation. Sporulation is an exquisitely choreographed event, requiring the synchronous compaction and segregation of chromosomes, such that the resulting ParB-oriC complexes are distributed at regularly spaced intervals along the sporogenic hyphae (Jakimowicz et al. 2005, 2007). At the same time, these multigenomic hyphae are subdivided into 10–50 unigenomic spore compartments (Fig. 1F). These processes require the upregulation of genes involved in chromosome segregation (parAB) (Jakimowicz et al. 2006), chromosome compaction (multiple nucleoid-associated proteins) and cell division (e.g. ftsZ) (Flärdh et al. 2000). FtsZ initiates cell division, forming an array of regularly spaced rings. The characterized streptomycete cell division machinery consists of a limited number of conserved cell division proteins, including FtsZ, FtsW and cell wall synthesizing enzymes (e.g. PBPs, FtsI); however, in contrast to unicellular bacteria, most of these proteins were dispensable for viability (although not for sporulation) (Flärdh and Buttner 2009; McCormick and Flärdh 2012). Moreover, the streptomycetes lack homologs of proteins involved in the regulation of septum positioning found in other bacteria, such as SlmA, Noc or Min (Wu and Errington 2004; Bernhardt and de Boer 2005; Lutkenhaus 2007). Instead, the positioning of FtsZ in sporulating hyphae is positively controlled by the Streptomyces-specific proteins SsgA and SsgB (Willemse et al. 2011; Jakimowicz and van Wezel 2012; McCormick and Flärdh 2012).
The mechanisms governing cell growth and chromosome dynamics during exploration are less well understood compared with vegetative and reproductive growth, and what features are conserved or required during this developmental stage remains an open question (Jones et al. 2017; Jones and Elliot 2018).
The onset of secondary metabolism, and the production of antibiotics and other natural products, are temporally and genetically coupled to reproductive growth (Hallam, Malpartida and Hopwood 1988). This metabolic transition does, however, tend to be spatially segregated from the reproductive cells, being confined to the vegetative hyphae. Increasingly, it appears that sporulation and secondary metabolism are both profoundly impacted by the activity of nucleoid-associated proteins, and other DNA-processing enzymes. Whether and how secondary metabolism is affected during the exploration phase of development remains to be determined.
CHROMOSOME ORGANIZATION DURING STREPTOMYCES VEGETATIVE GROWTH
The filamentous nature of Streptomyces vegetative hyphae and the polar tip/apical growth of these hyphal filaments mean that chromosome segregation and organization in these bacteria are subject to different constraints compared with most other bacteria. The large, linear nature of the Streptomyces chromosomes further expands the possible configurations that could be adopted by these molecules, compared with a circular chromosome of equivalent size. During vegetative growth, the multiple chromosomes are not obviously separated and remain uncondensed, occupying most of the hyphal cell volume (Kwak and Kendrick 1996). The development of fluorescence in situ hybridization (FISH) techniques and fluorescent reporter–operator systems (FROS) have made it possible to localize oriC and ter, and to follow the localization of segrosomes and replisomes (Yang and Losick 2001; Jakimowicz et al. 2005; Ruban-Ośmiałowska et al. 2006; Wolánski et al. 2011).
A distinctive feature of chromosome positioning during Streptomyces vegetative growth is the anchoring of the oriC of the apical chromosome to the growing hyphal tip. During spore germination, chromosome replication initiates before germ tube emergence. There is often more than one replisome detectable in germinating spores, suggesting that intensive chromosome replication may be required to ensure there are sufficient copies of the chromosome to populate the emerging hyphal tube(s) (Ruban-Ośmiałowska et al. 2006; Wolánski et al. 2011). The first chromosome is translocated into the growing hyphal filament once the germ tube is ∼2 μm long. This chromosome then remains in close proximity to the hyphal tip throughout growth (Kois-Ostrowska et al. 2016). In contrast to the precise positioning of the apical chromosome, the subapical copies exhibit more flexible distribution along the length of the hyphal filament, although they too follow the extending tip (Yang and Losick 2001; Kois-Ostrowska et al. 2016).
Notably, in Streptomyces hyphae, the oriC of the apical chromosome is positioned at the edge of the nucleoid, giving it a configuration similar to the polarly organized chromosomes of other bacteria like Caulobacter and Vibrio (Ebersbach et al. 2008; David et al. 2014; Kois-Ostrowska et al. 2016). Whether the subapical chromosomes are similarly oriented is not yet known. Using FISH, the ends of the linear chromosomes were found to co-localize with each other, but not with the oriC region (Yang and Losick 2001). This co-localization is assumed to be due to interaction of ter-bound proteins like Tap (telomere-associated protein) and/or Tpg (terminal protein gene) (Bao and Cohen 2003). Tap and Tpg maintain the integrity of the Streptomyces chromosomal termini, ensuring they are properly replicated by recruiting PolA (DNA polymerase) and TopA (topoisomerase) to the terminal telomeres (Bao and Cohen 2004). It is interesting to note, however, that Tap and Tpg are not universally conserved in the streptomycetes (see Table 2). Loss of Tap in S. coelicolor and S. lividans leads to DNA rearrangements, including chromosome circularization, which, surprisingly, does not visibly impair growth (Yang et al. 2002, 2013, 2017). Streptomyces genomes encode topoisomerase IV (encoded by parC and parE), which is primarily involved in the progression of replication forks and the decatenation of newly replicated DNA, and it is this decatenase activity that has the potential to support the separation of circular chromosomes, like those formed in the absence of Tap (Huang et al. 2013). Why Streptomyces chromosomes are linear remains a mystery, but this configuration presumably affords some fitness advantages over the more conventional circular orientation (Chen et al. 2002; Kirby 2011; Hopwood 2019).
Table 2.
Streptomyces proteins that contribute to chromosome dynamics or are abundant nucleoid-associated proteins.
| Protein name/S. coelicolor A3(2) gene number (Streptomyces venezuelaea gene number) | Predicted DNA-binding domain & sequence preference | Contribution to chromosome dynamics (experimentally determined) | Cell cycle stage when active (based on mutant phenotype or transcript/protein abundance) | Mutant effect on spore formation and resistance? | Reference |
|---|---|---|---|---|---|
| BldD/sco1489 (vnz_0 5285) | Yes; specific motif | Unknown | Vegetative growth (transcription) | Accelerated sporulation (no aerial hyphae formation) | (Elliot et al. 1998; Tschowri et al. 2014) |
| Crp/sco3571 (vnz_16 460) | Yes; specific motif | Unknown | Constitutive (protein abundance) | Accelerated sporulation | (Derouaux et al. 2004; Gao et al. 2012; Bradshaw, Saalbach and McArthur 2013) |
| DdbA/sco2075 (vnz_0 8485) | Yes; specificity unknown | Nucleoid condensation and nucleoid structure | Sporulation (transcript levels) | Defects in chromosome compaction and spore stress resistance | (Aldridge et al. 2013; Gehrke et al. 2019) |
| DpsA/sco0596 (n/a) | Yes; specificity unknown | Osmotic stress, heat shock | Sporulation (phenotype and immunoblotting); inducible in response to osmotic shock | Defects in chromosome segregation; heterogeneous spore sizes | (Facey et al. 2009) |
| DpsB/sco5756 (n/a) | Yes; specificity unknown | Nucleoid condensation (negative effect) | Sporulation (phenotype; low level constitutive expression determined via immunoblotting) | Shorter spores with more compact chromosomes | (Facey et al. 2009) |
| DpsC/sco1050 (vnz_34 055) | Yes; specificity unknown | Nucleoid condensation (negative effect) | Sporulation (phenotype; constitutive expression via immunoblotting); inducible at high temperatures | Shorter spores with more compact chromosomes | (Facey et al. 2009) |
| FtsK/sco5750 (vnz_26 720) | Yes; specificity unknown | Chromosome stability | Sporulation (reporter fusions) | Chromosome segregation defects | (Wang et al. 2007; Dedrick, Wildschutte and McCormick 2009) |
| GyrBA/sco3874–73 (vna17955–50) | Yes; specificity unknown | Replication and supercoiling | All (constitutive) | Predicted to be essential | n/a |
| HupA/sco2950 (vnz_13 325) | Yes; specificity unknown | Unknown | Vegetative growth (transcription) | Unknown | (Salerno et al. 2009) |
| HupS/sco5556 (vnz_25 950) | Yes; specificity unknown | Nucleoid compaction during sporulation | Sporulation (mutant phenotype and transcription) | Reduced pigmentation; reduced heat resistance | (Salerno et al. 2009) |
| Lsr2/sco3375 (vnz_15 890) | Yes; AT-rich sequences | Unknown | Onset of secondary metabolism (aerial hyphae formation) | Delayed sporulation | (Gehrke et al. 2019) |
| ParA/sco3886 (vnz_18 010) | Yes; non-specific binding predicted | Chromosome segregation | Vegetative (translocation into branches); sporulation (mutant phenotype and transcription) | Defects in chromosome segregation | (Jakimowicz et al. 2007) |
| ParB/sco3887 (vnz_18 015) | Yes; parS sequences | oriC organization and chromosome segregation | Vegetative (translocation into branches); sporulation (mutant phenotype and transcription) | Defects in chromosome segregation | (Kim et al. 2000; Jakimowicz et al. 2005) |
| SepG or YlmG/sco2078 (vnz_0 8500) | No (effects are likely indirect) | Nucleoid organization during sporulation | Sporulation (mutant phenotype) | Few spore chains; heterogeneous spore sizes | (Zhang et al. 2016) |
| SmeA-SffA/sco1415–16 (vnz_0 4885–90) | Unknown; unknown | Chromosome compaction and segregation | Sporulation | Defects in chromosome segregation; heterogeneous spore sizes; reduced heat resistance | (Ausmees et al. 2007) |
| sIHF/sco1480 (vnz_0 5240) | Yes; non-specific | Nucleoid compaction during sporulation | All (constitutive) | Delayed and reduced sporulation; defects in chromosome segregation; heterogeneous spore sizes | (Swiercz et al. 2013) |
| SMC + ScpAB/sco5577 + sco1770–69 (vnz_26 075 + vnz_06875–70) | Yes; specificity not tested | Chromosome compaction | Sporulation | Defects in chromosome segregation | (Dedrick, Wildschutte and McCormick 2009; Kois et al. 2009) |
| SsbB/sco2683 (vnz_12 100) | Yes; single-stranded DNA | Chromosome segregation | Sporulation (mutant phenotype) | Defects in chromosome segregation; heterogeneous spore sizes; delayed sporulation | (Paradzik et al. 2013) |
| Tap/sco7733 (n/a) | Yes; single-stranded telomeric DNA | Maintenance of telomeres and chromosome linearity | Unknown | Unknown | (Bao and Cohen 2003; Yang et al. 2017) |
| TopA/sco3543 (vnz_16 300) | Yes; non-specific but with AT-preference | DNA relaxation and chromosome decompaction; telomere maintenance | All (constitutive) | Essential gene; no sporulation in TopA-depletion strain | (Bao and Cohen 2004; Szafran et al. 2013, 2014) |
| Tpg/sco7734 (n/a) | Yes; non-specific (both single and double stranded DNA) | Maintenance of telomeres and chromosome linearity | Unknown | Unknown | (Bao and Cohen 2003; Yang et al. 2013) |
Streptomyces venezuelae strain NRRL B-65442.
The multigenomic nature of Streptomyces hyphal compartments raises the question of whether chromosome segregation is important during vegetative growth. Streptomyces genomes encode a typical ParABS segregation system in which the parAB genes are co-transcribed. Two promoters direct the expression of the parAB operon: one promoter is preferentially active during vegetative growth, while the second is active during aerial hyphae formation/sporulation (Jakimowicz et al. 2006). ParB binds multiple parS sites, which flank the oriC and are found throughout a region spanning ∼500 kb (Jakimowicz, Chater and Zakrzewska-Czerwińska 2002; Donczew et al. 2016). The number of ParB-binding sites in Streptomyces genomes exceeds that of most other bacteria. This increased concentration of parS sequences may be related to the relatively large size (∼8–10 Mb) of the Streptomyces chromosome. Alternatively, it may reflect the need for greater compaction due to the linear organization of Streptomyces chromosomes, or it may be related to the multigenomic nature of Streptomyces hyphal filaments. It is worth noting that the unigenomic bacterium Myxococcus xanthus has similar numbers of parS sequences on its large (∼9 Mb) circular chromosome (Harms et al. 2013; Iniesta 2014), suggesting that chromosome size may be the strongest driver of parS sequence number.
In vegetatively growing cells, all chromosomal oriC regions appear to be decorated with ParB (Kois-Ostrowska et al. 2016). Unexpectedly, ParB's binding partner, the ATPase ParA (Jakimowicz et al. 2007), is found predominantly at the hyphal tips, courtesy of its interaction with the Scy protein, an integral member of the tip polarisome complex (Walshaw, Gillespie and Kelemen 2010; Holmes et al. 2013). The polar positioning of ParA is necessary for anchoring the oriC of the apical chromosome to the tip: loss of ParA, or disruption of its interaction with either ParB or Scy, leads to impaired localization of the tip-associated oriC (Kois-Ostrowska et al. 2016). The interaction of ParA with polar proteins is reminiscent of the situation in C. crescentus, V. cholera and Mycobacterium tuberculosis, where ParA associates with dedicated polar tip proteins (TipN/PopZ, HpbP and DivIVA, respectively) (Ptacin et al. 2010, 2014; Ginda et al. 2013).
Streptomyces vegetative hyphae are characterized by the frequent initiation of branching, with continued growth of the new hyphal branch depending on the successful translocation of a chromosome into that growing branch (Kois-Ostrowska et al. 2016). Shortly after a new branch emerges, it is populated with a chromosome from the hyphal stem. ParA and ParB are needed to capture the chromosome at the base of the new branch, and promote its translocation into the emerging branch. The requirement for ParAB in populating new branches with chromosomes could explain their constitutive production during vegetative growth, despite no obvious need for chromosome segregation. It is worth noting, however, that mutations in parAB have only subtle phenotypic effects in vegetatively growing S. coelicolor (Kois-Ostrowska et al. 2016), at least under laboratory conditions. Whether these effects would have a significant fitness cost compared with wild-type strains, or during growth in the soil, remains to be determined.
Labeling of replisomes using DnaN fluorescent protein fusion has revealed that replication of the multiple chromosomes within the vegetative hyphal compartments is asynchronous (Ruban-Ośmiałowska et al. 2006). Moreover, the replicative activity differs between individual hyphal filaments. During replication, newly duplicated oriCs are bound by ParB; however, their separation efficiency depends on their position within the vegetative hyphae. While the subapical segrosomes seem to be separated passively and slowly, newly replicated oriCs associated with the apical chromosome are separated promptly after their duplication (Kois-Ostrowska et al. 2016). The efficiency of segrosome separation depends on the presence of apically localized ParA. Since ParA anchors one of the newly replicated oriCs at the tip, the simple act of hyphal tip extension seems to contribute to the separation of origins.
Chromosomes within the vegetative hyphae are not visibly compacted and occupy the majority of the intracellular space (Kwak and Kendrick 1996). This apparent loose, relaxed conformation needs to be maintained by DNA-organizing proteins, including topoisomerases and NAPs. Streptomyces coelicolor possesses a single essential DNA relaxase, named TopA, which is a highly processive enzyme (Szafran et al. 2013, 2014). Reducing TopA levels strongly increases chromosome negative supercoiling, inhibits growth rate, blocks sporulation and affects secondary metabolism (Szafran et al. 2013). Moreover, TopA depletion severely affects chromosome distribution in vegetative hyphal compartments, inhibiting the separation of ParB complexes and impeding oriC tip anchorage (Strzalka et al. 2017). TopA-depleted strains grow very slowly, and this may be due in part to the defects in chromosome distribution during vegetative growth. TopA depletion also results in pronounced transcriptional changes, including altered expression of nucleoid organizing protein-encoding genes (Szafran et al. 2019), suggesting that TopA activity is intertwined with that of NAPs, in maintaining appropriate chromosome topology.
Many NAPs are produced during Streptomyces vegetative growth (Table 2). These include the two HU homologs HupA and HupS, alongside sIHF, Dps-like proteins and Lsr2 (Facey et al. 2009; Salerno et al. 2009; Swiercz et al. 2013; Gehrke et al. 2019). Among these, HupA, Lsr2 and sIHF are the most abundant in S. coelicolor vegetative hyphae (Bradshaw, Saalbach and McArthur 2013; Szafran et al. 2019). To date, the role of HupA in Streptomyces growth is poorly understood, although in S. coelicolor, the hupA gene is transcribed most highly during vegetative growth (Szafran et al. 2019). In E. coli, HU proteins promote global organization of the nucleoid and facilitate chromosome segregation (Lioy et al. 2018). In S. lividans, a hupA deletion slows vegetative growth, but its effects on chromosome dynamics have not yet been probed (Yokoyama et al. 2001). Interestingly, expression of hupA is upregulated in response to increased negative supercoiling (Szafran et al. 2019). Like HupA, sIHF is produced constitutively during the Streptomyces life cycle, and is expressed most highly during vegetative growth, although its effects are most pronounced during reproductive growth (Bradshaw, Saalbach and McArthur 2013; Swiercz et al. 2013). In contrast to HupA and sIHF, loss of Lsr2 and Dps homologs did not visibly impair S. coelicolor vegetative growth (Fig. 1C), although sIHF and dps mutations decreased tolerance to stress conditions (Facey et al. 2009; Salerno et al. 2009; Gehrke et al. 2019). Similar observations have been made for E. coli and M. smegmatis, where strains with mutations in NAP-encoding genes were able to maintain their overall nucleoid architecture during growth under optimal conditions, but these mutant strains were more sensitive to stress than their wild-type parents (Dillon and Dorman 2010; Bartek et al. 2014; Holowka et al. 2017). The relatively minor growth effects associated with the loss of individual NAPs in Streptomyces, and other bacteria, suggest these proteins may share some level of functional redundancy.
CHROMOSOME DYNAMICS DURING AERIAL GROWTH AND SPORULATION
Streptomyces strains sporulate in response to nutrient limitation and stressful growth conditions. The vast majority of Streptomyces species can sporulate during growth on solid culture, but only a handful of tested species (e.g.S. venezuelae, S. griseus, S. albus) are capable of sporulating in liquid culture (Daza et al. 1989). Historically, the best-studied Streptomyces species has been S. coelicolor, which sporulates only during growth on solid medium. This has made it challenging to follow chromosome dynamics in real time, and thus, most information on chromosome organization during sporulation has come from analyses of still microscopy images. Recently, S. venezuelae has been developed for cell biology studies, and its ability to sporulate in liquid culture is enabling the direct microscopic observation of sporogenic hyphal development and the associated chromosome dynamics (Donczew et al. 2016; Schlimpert, Flärdh and Buttner 2016).
Before the onset of sporulation, the aerial or sporogenic hyphae rapidly elongate. This occurs in parallel with intensive DNA replication (Ruban-Ośmiałowska et al. 2006), yielding up to 50 chromosomes per sporogenic hyphal compartment (Fig. 1F and G). This rapid replication requires enhanced topoisomerase activity. Streptomyces (and their relatives) have unusually processive TopA enzymes, and this processivity appears to be related to a series of C-terminal Lys repeats that stabilize TopA−DNA interactions (Strzałka et al. 2017; Szafran, Strzałka and Jakimowicz 2020). While TopA levels are constant throughout the S. coelicolor life cycle (Szafran et al. 2013), depleting TopA completely blocks sporulation; however, lowering its processivity (by deleting Lys repeats) leads to shorter aerial compartments and shorter spore chains (Szafran et al. 2013; Strzalka et al. 2017), suggesting that high processivity is critical for maximal sporulation.
As aerial hyphae elongate and chromosome replication ramps up, ParAB and FtsZ production levels begin to rise. Expression of their associated genes depends on the aerial hyphae-specific regulators WhiA and WhiB (although only ftsZ is a direct target of these regulators) (Flärdh et al. 2000; Jakimowicz et al. 2006; Bush et al. 2013, 2016). Within these aerial compartments, ParA functions to both control hyphal elongation, and promote the uniform distribution of chromosomes down the length of the hyphae (Ditkowski et al. 2013; Donczew et al. 2016).
The ParA-mediated cessation of aerial growth is accompanied by the synchronous assembly of FtsZ in a ladder-like configuration extending the length of the aerial filament, delineating future spore compartments (Donczew et al. 2016). At the same time, ParB–DNA nucleoprotein complexes assemble, and ParA/B activity positions the oriC regions of chromosomes between FtsZ rings, such that each spore compartment inherits a single chromosome. Loss of parA or parB leads to increased numbers of anucleate spores (Kim et al. 2000; Jakimowicz et al. 2007), and further impacts FtsZ localization and spore septum positioning, suggesting a tight coordination of chromosome segregation and cell division during Streptomyces sporulation (Jakimowicz et al. 2007; Donczew et al. 2016). In C. crescentus and Rhodobacter spheroides, chromosome segregation is coupled to cell division through the interaction of ParB with MipZ, an oscillating protein that controls septum placement (Mohl and Gober 1997; Thanbichler and Shapiro 2006; Dubarry et al. 2019). Although, transmission electron micrographs depicting chromosomes spanning the space between encroaching cell division septa (e.g. Flärdh 2003b) suggest that unlike many other bacteria, Streptomyces lack an obvious nucleoid-occlusion system, the rigorous coordination between chromosome segregation and septum positioning indicates the existence of a dedicated control mechanism. How this control ties into the positive regulation of Z-ring formation by SsgA and SsgB remains to be determined. SepG represents an intriguing candidate for such a role: it is a membrane-associated protein that does not bind DNA directly, but strongly affects sporogenic nucleoid shape and influences SsgB localization (Zhang et al. 2016). Moreover, Streptomyces employ DNA pumps like FtsK and SffA, which appear to function to clear chromosomal DNA away from the closing septum (Ausmees et al. 2007; Sepulveda, Vogelmann and Muth 2011). Although many of the main players involved in cell division and chromosome segregation have been identified in Streptomyces, how their activities are coordinated to ensure appropriate synchronization requires further study.
The uniform distribution of ParB–DNA complexes within the sporogenic hyphae is abolished in TopA-depleted strains (Szafran et al. 2013). This inability to effectively segregate chromosomes has been proposed to explain the inhibition of cell division and overall spore formation observed for a TopA-depleted strain. The formation of the architecturally complex segrosome is expected to create topological tension within the oriC region, and resolution of this would require the relaxation activity of TopA activity. This notion is supported by the observations in S. coelicolor that TopA is recruited to the vicinity of ParB-occupied parS sites, and that parB deletion can partially suppress the sporulation defects associated with TopA depletion (Szafran et al. 2013). Since bacterial TopAs are recruited to single-stranded DNA (ssDNA) (Li, Mondragón and DiGate 2001), it has been suggested that segrosome assembly may generate ssDNA. Consistent with this proposal is the discovery that the single-stranded binding protein (SsbB) is required for chromosome segregation during S. coelicolor sporulation: the ssbB null mutant exhibits delayed sporulation and forms anucleate spores (Paradzik et al. 2013). Whether SsbB and TopA cooperate to alleviate the topological tension resulting from ParB-driven segrosome assembly during Streptomyces sporulation awaits future investigation.
The maturation of sporogenic hyphae is accompanied by a global rearrangement of Streptomyces nucleoid architecture—from uncondensed chromosomes during the initial growth of sporogenic/aerial hyphae, to highly compacted chromosomes during cell division and DNA segregation into pre-spore compartments (Donczew et al. 2016). In bacteria, the chromosome compacting activity of topoisomerases is coordinated with the activity of condensins (SMC/MukBEF) and nucleoid-associated proteins (Nolivos et al. 2016; Lioy et al. 2018). Surprisingly, deleting smc in S. coelicolor had only mild effects on DNA compaction and chromosome segregation (Dedrick, Wildschutte and McCormick 2009; Kois et al. 2009). Instead, chromosome defects were much more pronounced when the smc deletion was combined with either parB or ftsK deletions (Dedrick, Wildschutte and McCormick 2009), where ParB is part of the central chromosome segregation machinery, and FtsK functions as a translocase that pumps DNA out of the closing septum (Ausmees et al. 2007; Bigot et al. 2007). This suggests that chromosome compaction and segregation are driven by proteins sharing overlapping or redundant functions.
Beyond effective segregation, chromosomes must also be compacted during sporulation in order to effectively fit into the spore compartment, and to maximize resistance to DNA damage. A number of NAP-encoding genes are specifically upregulated during sporulation (Table 2), while other constitutively expressed ones have important functions during sporulation. Among them, DpsA, HupS and DdbA are key players, at least in S. coelicolor (Table 2). Each of their genes is expressed at low levels during vegetative and pre-sporogenic aerial growth, with their transcript levels rising dramatically during sporulation (Facey et al. 2009; Salerno et al. 2009; Aldridge et al. 2013). In the case of dpsA, its expression could also be induced during vegetative growth in response to osmotic stress, suggesting not only a developmental function for its gene product, but also a role in responding to environmental stresses. Streptomyces coelicolor dpsA mutants display variation in spore size and nucleoid volume. dpsA deletion further results in increased numbers of spores containing two or more nucleoids, implicating DpsA in chromosome segregation (Facey et al. 2009). It is worth noting, however, that DpsA is not broadly conserved in Streptomyces species (Table 2); whether other Dps proteins are able to substitute for it beyond S. coelicolor and its relatives has not yet been tested. In contrast, ddbA is highly conserved, and its deletion in S. coelicolor affects both chromosome compaction and spore resistance to various stresses, including osmotic and oxidative stress (Aldridge et al. 2013). Similar phenotypes were observed for a hupS mutant in S. coelicolor, which displayed reduced chromosome compaction, increased nucleoid size and decreased spore resistance to heat stress (Salerno et al. 2009). HupS is an HU-like protein, and like mycobacterial HupB and actinobacterial TopA (and eukaryotic histones), it possesses a C-terminal domain enriched in lysine repeats (Holowka et al. 2017; Szafran, Strzałka and Jakimowicz 2020); this Lys-rich repeat region is absent from the vegetative-specific HU-like protein HupA. Interestingly, an equivalent histone-like domain was also identified for DdbA within its N-terminus; truncation of DdbA at its N-terminus abolishes protein interaction with DNA (Aldridge et al. 2013).
Unlike DdbA, HupS and the Dps family of proteins, sIHF is an actinobacterial-specific nucleoid-associated protein that is expressed throughout growth, and functions to promote nucleoid condensation during sporulation (Swiercz et al. 2013; Nanji et al. 2019). Unlike its ortholog from Mycobacterium (mIHF) (Pedulla and Hatfull 1998; Odermatt et al. 2018), sIHF is not essential for S. coelicolor viability. Instead, its loss leads to reduced sporulation and aberrant nucleoid compaction (Swiercz et al. 2013). Unusually for a nucleoid-associated protein, sIHF functions as a monomer, and while it is only ∼100 aa in size, it has multiple DNA-binding faces. The crystal structure of sIHF revealed the presence of two DNA-binding surfaces capable of bridging two DNA duplexes, while subsequent small angle X-ray scattering structures revealed a third DNA-binding site (Nanji et al. 2019). In vitro experiments have revealed that sIHF binding to DNA can inhibit the DNA relaxation activity of TopA, and that it can act further to restrain negative supercoils (Swiercz et al. 2013; Nanji et al. 2019).
Our understanding of DNA organization during sporulation remains incomplete, as does our understanding of the interplay between the different chromosome-organizing proteins. Loss of any individual nucleoid-associated protein has a modest effect on the chromosome architecture during sporulation, suggesting that there is considerable functional redundancy shared between these proteins. The absence of individual nucleoid-associated proteins can, however, impact spore resistance to environmental stress, and can influence gene expression throughout the Streptomyces life cycle.
THE EFFECT OF CHROMOSOME-ORGANIZING PROTEINS ON GENE REGULATION AND CONTROL OF SECONDARY METABOLISM
In addition to playing fundamental roles in Streptomyces growth and development, chromosome-organizing proteins can also profoundly influence secondary metabolism, and make significant contributions to gene regulation. Recent work has revealed that Lsr2, an H-NS-like dimeric protein, functions as a major regulator of gene expression in S. venezuelae (Gehrke et al. 2019). Members of the Lsr2/H-NS family of proteins bind to AT-rich sequences and typically repress gene expression by either bridging DNA segments and trapping RNA polymerase, or polymerizing along the DNA and forming a rigid filament that is transcriptionally inactive (Dame 2005; Gordon et al. 2010; van der Valk et al. 2017). Coupling RNA-sequencing (RNA-seq) with the mapping of Lsr2-binding sites using chromatin immunoprecipitation and deep sequencing (ChIP-seq) led to the discovery that Lsr2 serves as a metabolic gatekeeper, directly repressing the expression of genes in the majority of secondary metabolic clusters in S. venezuelae (Gehrke et al. 2019). In contrast to Lsr2’s repressive effects, altering chromosome topology by either inhibiting gyrase or decreasing TopA levels in vegetatively growing S. coelicolor led to the discovery that changes in chromosome supercoiling also function as positive regulators of gene transcription, inducing the expression of several antibiotic biosynthetic clusters, among many other genes (Szafran et al. 2013, 2019). Indeed, long-term TopA depletion had pleiotropic effects and led to transcriptional changes in at least 7% of S. coelicolor genes. sIHF also impacts antibiotic production in a growth medium-dependent manner, although how its regulatory effects are exerted is currently unclear (Yang et al. 2012; Swiercz et al. 2013). On some media types, an sIHF mutant exhibits less antibiotic production than its wild-type parent, while on other media types, it overproduces antibiotics. It is conceivable that altered chromosomal supercoiling contributes to this differential antibiotic production by the sIHF mutant (as sIHF can inhibit TopA activity in vitro) (Swiercz et al. 2013), in addition to any direct regulatory effects stemming from its DNA binding.
Increasingly, the distinction between NAPs and transcription factors is becoming less clear (Dorman et al.2020), as is exemplified by two classical transcription regulators, Crp and BldC. Crp is conserved in many bacteria, and is one of the best-studied transcription factors. It is also among the most abundant DNA-binding proteins in S. coelicolor (Bradshaw, Saalbach and McArthur 2013), and is known to bend DNA when it binds its consensus sequence, thus remodeling the local DNA architecture; these are characteristics typically ascribed to NAPs. In S. coelicolor, Crp was experimentally determined to have ∼400 binding sites, and to impact the expression of similar numbers of genes (Gao et al. 2012). Like Lsr2, Crp appears to have a central role in governing secondary metabolism, only its effect is predominantly one of activation, in contrast to the repressive effects seen for Lsr2. More recently, the MerR-family transcription factor BldC has also been discovered to have NAP-like properties. This small regulator binds to DNA with flexible sequence specificity, and associates with DNA in a head-to-tail configuration that leads to both the formation of nucleoprotein filaments and distortion of the bound DNA (Bush et al. 2019). Again, the impact of BldC on DNA topology would suggest it functions as a nucleoid-associated protein. ChIP-seq experiments have revealed BldC binds to >350 sites throughout the S. venezuelae chromosome, while RNA-seq experiments revealed its regulatory effects to be both positive and negative. Unlike Crp and Lsr2, however, BldC has little effect on secondary metabolism, and instead has profound impacts on classical development, influencing the expression of many genes needed for growth (e.g.divIVA), cell division (e.g.ftsZ) and chromosome segregation and compaction (e.g.hupS, sepG and sffA) (Bush et al. 2019).
Manipulating the activities of Lsr2 and Crp has proven to be productive avenue for stimulating the expression and production of ‘cryptic’ secondary metabolites, and it is conceivable that modulating the function of other NAPs, and/or generally altering chromosome architecture or local domain structure, may yield new activation strategies for many of the uncharacterized biosynthetic clusters encoded in the genomes of Streptomyces spp. Proteomic analysis of the S. coelicolor nucleoid (Bradshaw, Saalbach and McArthur 2013) revealed many of the expected NAPs (HupA, HupS, sIHF, Lsr2, Crp). But alongside these were many uncharacterized proteins, as well as more conventional transcription factors like BldD and AfsQ1, where BldD is a well-studied transcription factor that controls both development and secondary metabolism in various streptomycetes (Elliot et al. 1998; den Hengst et al. 2010; Wang et al. 2013) and AfsQ1 is a classical response regulator that has antibiotic modulatory capabilities (Ishizuka et al. 1992; Daniel-Ivan et al. 2017). It is clear that we are lacking a comprehensive understanding of the factors capable of influencing chromosome structure in the streptomycetes, and as we begin to appreciate how these different proteins function, there will be a corresponding opportunity to shed light on new levels of secondary metabolic control.
Chromosome dynamics are intimately entwined with transcriptional regulation, and it will be important to consider proteins involved in chromosome organization, when defining any regulatory cascade of interest. Reciprocally, it will be critical to account for the architectural contributions made by ‘conventional’ transcription factors, when investigating the factors that influence chromosome structure.
CONCLUDING REMARKS
It is an exciting time to be probing chromosome dynamics and the diverse roles and functions of the proteins that influence chromosome compaction, segregation, organization, and transcription. The unique life cycle of Streptomyces provides an outstanding opportunity to explore chromosome dynamics and organization in diverse contexts, and investigations are revealing intriguing adaptations that allow these bacteria to thrive. An ambitious, long-term goal will be to achieve a systems-level understanding of how the activity of the diverse proteins summarized in Table 2 collectively functions to ensure chromosome integrity and functionality.
ACKNOWLEDGEMENTS
We would like to thank the Canadian Institutes of Health Research (MAE) and the Boris Family Foundation (MAE) for their support of this work. MJS and DJ acknowledge support from the National Science Centre, Poland: HARMONIA grant 2016/22/NZ1/00122 (MJS), SONATA grant 2018/31/D/NZ1/00287 (MJS) and OPUS grant 2018/31/B/NZ1/00614 (DJ).
Contributor Information
Marcin J Szafran, Laboratory of Molecular Microbiology, Faculty of Biotechnology, University of Wroclaw, 50-383 Wroclaw, Poland.
Dagmara Jakimowicz, Laboratory of Molecular Microbiology, Faculty of Biotechnology, University of Wroclaw, 50-383 Wroclaw, Poland.
Marie A Elliot, Department of Biology, Michael G. DeGroote Institute for Infectious Disease Research, McMaster University, Hamilton, ON, L8S 4K1, Canada.
Conflict of interest
The authors declare no conflict of interest.
REFERENCES
- Ahmed W, Menon S, Karthik PVDNB et al. Autoregulation of topoisomerase I expression by supercoiling sensitive transcription. Nucleic Acids Res. 2015, DOI: 10.1093/nar/gkv1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldridge M, Facey P, Francis L et al. A novel bifunctional histone protein in Streptomyces: a candidate for structural coupling between DNA conformation and transcription during development and stress? Nucleic Acids Res. 2013;41:4813–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausmees N, Wahlstedt H, Bagchi S et al. SmeA, a small membrane protein with multiple functions in Streptomyces sporulation including targeting of a SpoIIIE/FtsK-like protein to cell division septa. Mol Microbiol. 2007;65:1458–73. [DOI] [PubMed] [Google Scholar]
- Badrinarayanan A, Le TBK, Laub MT. Bacterial chromosome organization and segregation. Annu Rev Cell Dev Biol. 2015;31:171–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao K, Cohen SN. Recruitment of terminal protein to the ends of Streptomyces linear plasmids and chromosomes by a novel telomere-binding protein essential for linear DNA replication. Genes Dev. 2003;17:774–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao K, Cohen SN. Reverse transcriptase activity innate to DNA polymerase I and DNA topoisomerase I proteins of Streptomyces telomere complex. Proc Natl Acad Sci USA. 2004;101:14361–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartek IL, Woolhiser LK, Baughn AD et al. Mycobacterium tuberculosis Lsr2 is a global transcriptional regulator required for adaptation to changing oxygen levels and virulence. mBio. 2014;5:e01106–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Yehuda S, Rudner DZ, Losick R. RacA, a bacterial protein that anchors chromosomes to the cell poles. Science. 2003;299:532–6. [DOI] [PubMed] [Google Scholar]
- Bernhardt TG, de Boer PAJ. SlmA, a nucleoid-associated, FtsZ binding protein required for blocking septal ring assembly over chromosomes in E. coli. Mol Cell. 2005;18:555–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigot S, Sivanathan V, Possoz C et al. FtsK, a literate chromosome segregation machine. Mol Microbiol. 2007;64:1434–41. [DOI] [PubMed] [Google Scholar]
- Bradshaw E, Saalbach G, McArthur M. Proteomic survey of the Streptomyces coelicolor nucleoid. J Proteomics. 2013;83:37–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brochu J, Breton É-V, Drolet M. Supercoiling, R-loops, replication and the functions of bacterial type 1A topoisomerases. Genes. 2020;11, DOI: 10.3390/genes11030249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush MJ, Bibb MJ, Chandra G et al. Genes required for aerial growth, cell division, and chromosome segregation are targets of WhiA before sporulation in Streptomyces venezuelae. mBio. 2013;4:e00684–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush MJ, Chandra G, Al-Bassam MM et al. BldC delays entry into development to produce a sustained period of vegetative growth in Streptomyces venezuelae. mBio. 2019;10, DOI: 10.1128/mBio.02812-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush MJ, Chandra G, Bibb MJ et al. Genome-wide chromatin immunoprecipitation sequencing analysis shows that WhiB is a transcription factor that co-controls its regulon with WhiA to initiate developmental cell division in Streptomyces. mBio. 2016;7:e00523–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CW, Huang C-H, Lee H-H et al. Once the circle has been broken: dynamics and evolution of Streptomyces chromosomes. Trends Genet. 2002;18:522–9. [DOI] [PubMed] [Google Scholar]
- Dame RT, Rashid F-ZM, Grainger DC. Chromosome organization in bacteria: mechanistic insights into genome structure and function. Nat Rev Genet. 2019, DOI: 10.1038/s41576-019-0185-4. [DOI] [PubMed] [Google Scholar]
- Dame RT. The role of nucleoid-associated proteins in the organization and compaction of bacterial chromatin. Mol Microbiol. 2005;56:858–70. [DOI] [PubMed] [Google Scholar]
- Daniel-Ivad M, Hameed N, Tan S et al. An engineered allele of afsQ1 facilitates the discovery and investigation of cryptic natural products. ACS Chem Biol. 2017;12:628–34. [DOI] [PubMed] [Google Scholar]
- David A, Demarre G, Muresan L et al. The two cis-acting sites, parS1 and oriC1, contribute to the longitudinal organisation of Vibrio cholerae chromosome I. PLos Genet. 2014;10:e1004448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daza A, Martín JF, Dominguez A et al. Sporulation of several species of Streptomyces in submerged cultures after nutritional downshift. J Gen Microbiol. 1989;135:2483–91. [DOI] [PubMed] [Google Scholar]
- Dedrick RM, Wildschutte H, McCormick JR. Genetic interactions of smc, ftsK, and parB genes in Streptomyces coelicolor and their developmental genome segregation phenotypes. J Bacteriol. 2009;191:320–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Hengst CD, Tran NT, Bibb MJ et al. Genes essential for morphological development and antibiotic production in Streptomyces coelicolor are targets of BldD during vegetative growth. Mol Microbiol. 2010;78:361–79. [DOI] [PubMed] [Google Scholar]
- Derouaux A, Dehareng D, Lecocq E et al. Crp of Streptomyces coelicolor is the third transcription factor of the large CRP-FNR superfamily able to bind cAMP. Biochem Biophys Res Commun. 2004;325:983–90. [DOI] [PubMed] [Google Scholar]
- de Vries R. DNA condensation in bacteria: interplay between macromolecular crowding and nucleoid proteins. Biochimie. 2010;92:1715–21. [DOI] [PubMed] [Google Scholar]
- Dillon SC, Dorman CJ. Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat Rev Microbiol. 2010;8:185–95. [DOI] [PubMed] [Google Scholar]
- Ditkowski B, Holmes N, Rydzak J et al. Dynamic interplay of ParA with the polarity protein, Scy, coordinates the growth with chromosome segregation in Streptomyces coelicolor. Open Biol. 2013;3:130006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donczew M, Mackiewicz P, Wróbel A et al. ParA and ParB coordinate chromosome segregation with cell elongation and division during Streptomyces sporulation. Open Biol. 2016;6:150263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorman CJ, Schumacher MA, Bush MJ et al. When is a transcription factor a NAP? Curr Opin Microbiol. 2020;55:26–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorman CJ. Genome architecture and global gene regulation in bacteria: making progress towards a unified model? Nat Rev Microbiol. 2013;11:349–55. [DOI] [PubMed] [Google Scholar]
- Dubarry N, Willis CR, Ball G et al. In vivo imaging of the segregation of the 2 chromosomes and the cell division proteins of Rhodobacter sphaeroides reveals an unexpected role for MipZ. mBio. 2019;10, DOI: 10.1128/mBio.02515-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebersbach G, Briegel A, Jensen GJ et al. A self-associating protein critical for chromosome attachment, division, and polar organization in Caulobacter. Cell. 2008;134:956–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliot M, Damji F, Passantino R et al. The bldD gene of Streptomyces coelicolor A3(2): a regulatory gene involved in morphogenesis and antibiotic production. J Bacteriol. 1998;180:1549–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliot MA, Flärdh K. Streptomycete Spores. 2012, DOI: 10.1002/9780470015902.a0000308.pub2. [Google Scholar]
- Errington J. Septation and chromosome segregation during sporulation in Bacillus subtilis. Curr Opin Microbiol. 2001;4:660–6. [DOI] [PubMed] [Google Scholar]
- Espeli O, Mercier R, Boccard F. DNA dynamics vary according to macrodomain topography in the E. coli chromosome. Mol Microbiol. 2008;68:1418–27. [DOI] [PubMed] [Google Scholar]
- Espéli O, Boccard F. Organization of the Escherichia coli chromosome into macrodomains and its possible functional implications. J Struct Biol. 2006;156:304–10. [DOI] [PubMed] [Google Scholar]
- Facey PD, Hitchings MD, Saavedra-Garcia P et al. Streptomyces coelicolor Dps-like proteins: differential dual roles in response to stress during vegetative growth and in nucleoid condensation during reproductive cell division. Mol Microbiol. 2009;73:1186–202. [DOI] [PubMed] [Google Scholar]
- Feijoo-Siota L, Rama JLR, Sánchez-Pérez A et al. Considerations on bacterial nucleoids. Appl Microbiol Biotechnol. 2017;101:5591–602. [DOI] [PubMed] [Google Scholar]
- Ferrandiz M-J, Martin-Galiano AJ, Arnanz C et al. An increase in negative supercoiling in bacteria reveals topology-reacting gene clusters and a homeostatic response mediated by the DNA topoisomerase I gene. Nucleic Acids Res. 2016;44:7292–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrándiz M-J, Carreño D, Ayora S et al. HU of Streptococcus pneumoniae Is essential for the preservation of DNA supercoiling. Front Microbiol. 2018;9:493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flärdh K, Buttner MJ. Streptomyces morphogenetics: dissecting differentiation in a filamentous bacterium. Nat Rev Microbiol. 2009;7:36–49. [DOI] [PubMed] [Google Scholar]
- Flärdh K, Leibovitz E, Buttner MJ et al. Generation of a non-sporulating strain of Streptomyces coelicolor A3(2) by the manipulation of a developmentally controlled ftsZ promoter. Mol Microbiol. 2000;38:737–49. [DOI] [PubMed] [Google Scholar]
- Flärdh K, Richards DM, Hempel AM et al. Regulation of apical growth and hyphal branching in Streptomyces. Curr Opin Microbiol. 2012;15:737–43. [DOI] [PubMed] [Google Scholar]
- Flärdh K. Essential role of DivIVA in polar growth and morphogenesis in Streptomyces coelicolor A3(2). Mol Microbiol. 2003a;49:1523–36. [DOI] [PubMed] [Google Scholar]
- Flärdh K. Growth polarity and cell division in Streptomyces. Curr Opin Microbiol. 2003b;6:564–71. [DOI] [PubMed] [Google Scholar]
- Forterre P, Gribaldo S, Gadelle D et al. Origin and evolution of DNA topoisomerases. Biochimie. 2007;89:427–46. [DOI] [PubMed] [Google Scholar]
- Fröjd MJ, Flärdh K. Apical assemblies of intermediate filament-like protein FilP are highly dynamic and affect polar growth determinant DivIVA in Streptomyces venezuelae. Mol Microbiol. 2019;112:47–61. [DOI] [PubMed] [Google Scholar]
- Fuchino K, Bagchi S, Cantlay S et al. Dynamic gradients of an intermediate filament-like cytoskeleton are recruited by a polarity landmark during apical growth. Proc Natl Acad Sci USA. 2013;110:E1889–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C, null H, Mulder D et al. Crp is a global regulator of antibiotic production in Streptomyces. mBio. 2012;3, DOI: 10.1128/mBio.00407-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehrke EJ, Zhang X, Pimentel-Elardo SM et al. Silencing cryptic specialized metabolism in Streptomyces by the nucleoid-associated protein Lsr2. eLife. 2019;8, DOI: 10.7554/eLife.47691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Mallick B, Nagaraja V. Direct regulation of topoisomerase activity by a nucleoid-associated protein. Nucleic Acids Res. 2014;42:11156–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginda K, Bezulska M, Ziółkiewicz M et al. ParA of Mycobacterium smegmatis co-ordinates chromosome segregation with the cell cycle and interacts with the polar growth determinant DivIVA. Mol Microbiol. 2013;87:998–1012. [DOI] [PubMed] [Google Scholar]
- Gordon BRG, Li Y, Wang L et al. Lsr2 is a nucleoid-associated protein that targets AT-rich sequences and virulence genes in Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2010;107:5154–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham TGW, Wang X, Song D et al. ParB spreading requires DNA bridging. Genes Dev. 2014;28:1228–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray DI, Gooday GW, Prosser JI. Apical hyphal extension in Streptomyces coelicolor A3(2). J Gen Microbiol. 1990;136:1077–84. [DOI] [PubMed] [Google Scholar]
- Gruber S, Errington J. Recruitment of condensin to replication origin regions by ParB/SpoOJ promotes chromosome segregation in B. subtilis. Cell. 2009;137:685–96. [DOI] [PubMed] [Google Scholar]
- Hallam SE, Malpartida F, Hopwood DA. Nucleotide sequence, transcription and deduced function of a gene involved in polyketide antibiotic synthesis in Streptomyces coelicolor. Gene. 1988;74:305–20. [DOI] [PubMed] [Google Scholar]
- Hardy CD, Cozzarelli NR. A genetic selection for supercoiling mutants of Escherichia coli reveals proteins implicated in chromosome structure. Mol Microbiol. 2005;57:1636–52. [DOI] [PubMed] [Google Scholar]
- Harms A, Treuner-Lange A, Schumacher D et al. Tracking of chromosome and replisome dynamics in Myxococcus xanthus reveals a novel chromosome arrangement. PLos Genet. 2013;9:e1003802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayama R, Marians KJ. Physical and functional interaction between the condensin MukB and the decatenase topoisomerase IV in Escherichia coli. Proc Natl Acad Sci USA. 2010;107:18826–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hempel AM, Wang S, Letek M et al. Assemblies of DivIVA mark sites for hyphal branching and can establish new zones of cell wall growth in Streptomyces coelicolor. J Bacteriol. 2008;190:7579–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes NA, Walshaw J, Leggett RM et al. Coiled-coil protein Scy is a key component of a multiprotein assembly controlling polarized growth in Streptomyces. Proc Natl Acad Sci USA. 2013;110:E397–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holowka J, Trojanowski D, Ginda K et al. HupB Is a bacterial nucleoid-associated protein with an Indispensable eukaryotic-like tail. mBio. 2017;8, DOI: 10.1128/mBio.01272-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopwood DA. Highlights of Streptomyces genetics. Heredity. 2019;123:23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hołówka J, Trojanowski D, Janczak M et al. The origin of chromosomal replication Is asymmetrically positioned on the mycobacterial nucleoid, and the timing of its firing depends on HupB. J Bacteriol. 2018;200, DOI: 10.1128/JB.00044-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T-W, Hsu C-C, Yang H-Y et al. Topoisomerase IV is required for partitioning of circular chromosomes but not linear chromosomes in Streptomyces. Nucleic Acids Res. 2013;41:10403–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iniesta AA. ParABS system in chromosome partitioning in the bacterium Myxococcus xanthus. PLoS One. 2014;9:e86897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizuka H, Horinouchi S, Kieser HM et al. A putative two-component regulatory system involved in secondary metabolism in Streptomyces spp. J Bacteriol. 1992;174:7585–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakimowicz D, Chater K, Zakrzewska-Czerwínska J. The ParB protein of Streptomyces coelicolor A3(2) recognizes a cluster of parS sequences within the origin-proximal region of the linear chromosome. Mol Microbiol. 2002;45:1365–77. [DOI] [PubMed] [Google Scholar]
- Jakimowicz D, Gust B, Zakrzewska-Czerwinska J et al. Developmental-stage-specific assembly of ParB complexes in Streptomyces coelicolor hyphae. J Bacteriol. 2005;187:3572–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakimowicz D, Mouz S, Zakrzewska-Czerwinska J et al. Developmental control of a parAB promoter leads to formation of sporulation-associated ParB complexes in Streptomyces coelicolor. J Bacteriol. 2006;188:1710–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakimowicz D, van Wezel GP. Cell division and DNA segregation in Streptomyces: how to build a septum in the middle of nowhere? Mol Microbiol. 2012;85:393–404. [DOI] [PubMed] [Google Scholar]
- Jakimowicz D, Zydek P, Kois A et al. Alignment of multiple chromosomes along helical ParA scaffolding in sporulating Streptomyces hyphae. Mol Microbiol. 2007;65:625–41. [DOI] [PubMed] [Google Scholar]
- Jalal AS, Tran NT, Le TB. ParB spreading on DNA requires cytidine triphosphate in vitro. eLife. 2020;9, DOI: 10.7554/eLife.53515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SE, Elliot MA. “Exploring” the regulation of Streptomyces growth and development. Curr Opin Microbiol. 2018;42:25–30. [DOI] [PubMed] [Google Scholar]
- Jones SE, Ho L, Rees CA et al. Streptomyces exploration is triggered by fungal interactions and volatile signals. eLife. 2017;6, DOI: 10.7554/eLife.21738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawalek A, Wawrzyniak P, Bartosik AA et al. Rules and exceptions: the role of chromosomal ParB in DNA segregation and other cellular processes. Microorganisms. 2020;8, DOI: 10.3390/microorganisms8010105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Calcutt MJ, Schmidt FJ et al. Partitioning of the linear chromosome during sporulation of Streptomyces coelicolor A3(2) involves an oriC-linked parAB locus. J Bacteriol. 2000;182:1313–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby R. Chromosome diversity and similarity within the Actinomycetales. FEMS Microbiol Lett. 2011;319:1–10. [DOI] [PubMed] [Google Scholar]
- Kleckner N, Fisher JK, Stouf M et al. The bacterial nucleoid: nature, dynamics and sister segregation. Curr Opin Microbiol. 2014;22:127–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kois-Ostrowska A, Strzałka A, Lipietta N et al. Unique function of the bacterial chromosome segregation machinery in apically growing Streptomyces – targeting the chromosome to new hyphal tubes and its anchorage at the tips. PLos Genet. 2016;12:e1006488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kois A, Swiatek M, Jakimowicz D et al. SMC protein-dependent chromosome condensation during aerial hyphal development in Streptomyces. J Bacteriol. 2009;191:310–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak J, Kendrick KE. Bald mutants of Streptomyces griseus that prematurely undergo key events of sporulation. J Bacteriol. 1996;178:4643–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal A, Dhar A, Trostel A et al. Genome scale patterns of supercoiling in a bacterial chromosome. Nat Commun. 2016;7:11055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le TBK, Imakaev MV, Mirny LA et al. High-resolution mapping of the spatial organization of a bacterial chromosome. Science. 2013;342:731–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim HC, Surovtsev IV, Beltran BG et al. Evidence for a DNA-relay mechanism in ParABS-mediated chromosome segregation. eLife. 2014;3:e02758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin DC, Grossman AD. Identification and characterization of a bacterial chromosome partitioning site. Cell. 1998;92:675–85. [DOI] [PubMed] [Google Scholar]
- Lioy VS, Cournac A, Marbouty M et al. Multiscale structuring of the E. coli chromosome by nucleoid-associated and condensin proteins. Cell. 2018;172:771–83..e18. [DOI] [PubMed] [Google Scholar]
- Livny J, Yamaichi Y, Waldor MK. Distribution of centromere-like parS sites in bacteria: insights from comparative genomics. J Bacteriol. 2007;189:8693–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Mondragón A, DiGate RJ. The mechanism of type IA topoisomerase-mediated DNA topological transformations. Mol Cell. 2001;7:301–7. [DOI] [PubMed] [Google Scholar]
- Lutkenhaus J. Assembly dynamics of the bacterial MinCDE system and spatial regulation of the Z ring. Annu Rev Biochem. 2007;76:539–62. [DOI] [PubMed] [Google Scholar]
- Marko JF, De Los Rios P, Barducci A et al. DNA-segment-capture model for loop extrusion by structural maintenance of chromosome (SMC) protein complexes. Nucleic Acids Res. 2019;47:6956–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick JR, Flärdh K. Signals and regulators that govern Streptomyces development. FEMS Microbiol Rev. 2012;36:206–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick JR. Cell division is dispensable but not irrelevant in Streptomyces. Curr Opin Microbiol. 2009;12:689–98. [DOI] [PubMed] [Google Scholar]
- Mercier R, Petit M-A, Schbath S et al. The MatP/matS site-specific system organizes the terminus region of the E. coli chromosome into a macrodomain. Cell. 2008;135:475–85. [DOI] [PubMed] [Google Scholar]
- Minnen A, Attaiech L, Thon M et al. SMC is recruited to oriC by ParB and promotes chromosome segregation in Streptococcus pneumoniae. Mol Microbiol. 2011;81:676–88. [DOI] [PubMed] [Google Scholar]
- Mohl DA, Gober JW. Cell cycle-dependent polar localization of chromosome partitioning proteins in Caulobacter crescentus. Cell. 1997;88:675–84. [DOI] [PubMed] [Google Scholar]
- Murphy LD, Zimmerman SB. Condensation and cohesion of lambda DNA in cell extracts and other media: implications for the structure and function of DNA in prokaryotes. Biophys Chem. 1995;57:71–92. [DOI] [PubMed] [Google Scholar]
- Mäkelä J, Sherratt DJ. Organization of the Escherichia coli chromosome by a MukBEF axial core. Mol Cell. 2020, DOI: 10.1016/j.molcel.2020.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanji T, Gehrke EJ, Shen Y et al. Streptomyces IHF uses multiple interfaces to bind DNA. Biochim Biophys Acta Gen Subj. 2019;1863:129405. [DOI] [PubMed] [Google Scholar]
- Niki H, Hiraga S. Polar localization of the replication origin and terminus in Escherichia coli nucleoids during chromosome partitioning. Genes Dev. 1998;12:1036–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niki H, Yamaichi Y, Hiraga S. Dynamic organization of chromosomal DNA in Escherichia coli. Genes Dev. 2000;14:212–23. [PMC free article] [PubMed] [Google Scholar]
- Nolivos S, Upton AL, Badrinarayanan A et al. MatP regulates the coordinated action of topoisomerase IV and MukBEF in chromosome segregation. Nat Commun. 2016;7:10466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odermatt NT, Sala C, Benjak A et al. Essential nucleoid associated protein mIHF (Rv1388) controls virulence and housekeeping genes in Mycobacterium tuberculosis. Sci Rep. 2018;8:14214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osorio-Valeriano M, Altegoer F, Steinchen W et al. ParB-type DNA segregation proteins are CTP-dependent molecular switches. Cell. 2019;179:1512–24..e15. [DOI] [PubMed] [Google Scholar]
- Paradzik T, Ivic N, Filic Z et al. Structure-function relationships of two paralogous single-stranded DNA-binding proteins from Streptomyces coelicolor: implication of SsbB in chromosome segregation during sporulation. Nucleic Acids Res. 2013;41:3659–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedulla ML, Hatfull GF. Characterization of the mIHF gene of Mycobacterium smegmatis. J Bacteriol. 1998;180:5473–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postow L, Hardy CD, Arsuaga J et al. Topological domain structure of the Escherichia coli chromosome. Genes Dev. 2004;18:1766–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptacin JL, Gahlmann A, Bowman GR et al. Bacterial scaffold directs pole-specific centromere segregation. Proc Natl Acad Sci USA. 2014;111:E2046–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptacin JL, Lee SF, Garner EC et al. A spindle-like apparatus guides bacterial chromosome segregation. Nat Cell Biol. 2010;12:791–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricci DP, Melfi MD, Lasker K et al. Cell cycle progression in Caulobacter requires a nucleoid-associated protein with high AT sequence recognition. Proc Natl Acad Sci USA. 2016;113:E5952–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards DM, Hempel AM, Flärdh K et al. Mechanistic basis of branch-site selection in filamentous bacteria. PLOS Comput Biol. 2012;8:e1002423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruban-Ośmiałowska B, Jakimowicz D, Smulczyk-Krawczyszyn A et al. Replisome localization in vegetative and aerial hyphae of Streptomyces coelicolor. J Bacteriol. 2006;188:7311–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salerno P, Larsson J, Bucca G et al. One of the two genes encoding nucleoid-associated HU proteins in Streptomyces coelicolor is developmentally regulated and specifically involved in spore maturation. J Bacteriol. 2009;191:6489–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlimpert S, Flärdh K, Buttner M. Fluorescence time-lapse imaging of the complete S. venezuelae life cycle using a microfluidic device. J Vis Exp. 2016:53863, DOI: 10.3791/53863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepulveda E, Vogelmann J, Muth G. A septal chromosome segregator protein evolved into a conjugative DNA-translocator protein. Mob Genet Elem. 2011;1:225–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen BA, Landick R. Transcription of bacterial chromatin. J Mol Biol. 2019;431:4040–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soh Y-M, Davidson IF, Zamuner S et al. Self-organization of parS centromeres by the ParB CTP hydrolase. Science. 2019;366:1129–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song D, Rodrigues K, Graham TGW et al. A network of cis and trans interactions is required for ParB spreading. Nucleic Acids Res. 2017;45:7106–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strzalka A, Szafran MJ, Strick T et al. C-terminal lysine repeats in Streptomyces topoisomerase I stabilize the enzyme-DNA complex and confer high enzyme processivity. Nucleic Acids Res. 2017, DOI: 10.1093/nar/gkx827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan NL, Marquis KA, Rudner DZ. Recruitment of SMC by ParB-parS organizes the origin region and promotes efficient chromosome segregation. Cell. 2009;137:697–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surovtsev IV, Campos M, Jacobs-Wagner C. DNA-relay mechanism is sufficient to explain ParA-dependent intracellular transport and patterning of single and multiple cargos. Proc Natl Acad Sci USA. 2016;113:E7268–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiercz JP, Nanji T, Gloyd M et al. A novel nucleoid-associated protein specific to the actinobacteria. Nucleic Acids Res. 2013;41:4171–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szafran M, Skut P, Ditkowski B et al. Topoisomerase I (TopA) is recruited to ParB complexes and is required for proper chromosome organization during Streptomyces coelicolor sporulation. J Bacteriol. 2013;195:4445–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szafran MJ, Gongerowska M, Gutkowski P et al. The coordinated positive regulation of topoisomerase genes maintains topological homeostasis in Streptomyces coelicolor. J Bacteriol. 2016, DOI: 10.1128/JB.00530-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szafran MJ, Gongerowska M, Małecki T et al. Transcriptional response of Streptomyces coelicolor to rapid chromosome relaxation or long-term supercoiling imbalance. Front Microbiol. 2019;10:1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szafran MJ, Strick T, Strzałka A et al. A highly processive topoisomerase I: studies at the single-molecule level. Nucleic Acids Res. 2014;42:7935–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szafran MJ, Strzałka A, Jakimowicz D. A highly processive actinobacterial topoisomerase I – thoughts on Streptomyces’ demand for an enzyme with a unique C-terminal domain. Microbiol Read Engl. 2020;166:120–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanbichler M, Shapiro L. MipZ, a spatial regulator coordinating chromosome segregation with cell division in Caulobacter. Cell. 2006;126:147–62. [DOI] [PubMed] [Google Scholar]
- Tschowri N, Schumacher MA, Schlimpert S et al. Tetrameric c-di-GMP mediates effective transcription factor dimerization to control Streptomyces development. Cell. 2014;158:1136–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valens M, Penaud S, Rossignol M et al. Macrodomain organization of the Escherichia coli chromosome. EMBO J. 2004;23:4330–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valens M, Thiel A, Boccard F. The MaoP/maoS site-specific system organizes the ori region of the E. coli chromosome into a macrodomain. PLos Genet. 2016;12:e1006309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Valk RA, Vreede J, Qin L et al. Mechanism of environmentally driven conformational changes that modulate H-NS DNA-bridging activity. eLife. 2017;6, DOI: 10.7554/eLife.27369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vecchiarelli AG, Neuman KC, Mizuuchi K. A propagating ATPase gradient drives transport of surface-confined cellular cargo. Proc Natl Acad Sci USA. 2014;111:4880–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viollier PH, Thanbichler M, McGrath PT et al. Rapid and sequential movement of individual chromosomal loci to specific subcellular locations during bacterial DNA replication. Proc Natl Acad Sci USA. 2004;101:9257–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walshaw J, Gillespie MD, Kelemen GH. A novel coiled-coil repeat variant in a class of bacterial cytoskeletal proteins. J Struct Biol. 2010;170:202–15. [DOI] [PubMed] [Google Scholar]
- Wang L, Yu Y, He X et al. Role of an FtsK-like protein in genetic stability in Streptomyces coelicolor A3(2). J Bacteriol. 2007;189:2310–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Mast Y, Wang J et al. Identification of two-component system AfsQ1/Q2 regulon and its cross-regulation with GlnR in Streptomyces coelicolor. Mol Microbiol. 2013;87:30–48. [DOI] [PubMed] [Google Scholar]
- Wang X, Brandão HB, Le TBK et al. Bacillus subtilis SMC complexes juxtapose chromosome arms as they travel from origin to terminus. Science. 2017;355:524–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Montero Llopis P, Rudner DZ. Bacillus subtilis chromosome organization oscillates between two distinct patterns. Proc Natl Acad Sci USA. 2014;111:12877–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Montero Llopis P, Rudner DZ. Organization and segregation of bacterial chromosomes. Nat Rev Genet. 2013;14:191–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb CD, Teleman A, Gordon S et al. Bipolar localization of the replication origin regions of chromosomes in vegetative and sporulating cells of B. subtilis. Cell. 1997;88:667–74. [DOI] [PubMed] [Google Scholar]
- Willemse J, Borst JW, de Waal E et al. Positive control of cell division: FtsZ is recruited by SsgB during sporulation of Streptomyces. Genes Dev. 2011;25:89–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolánski M, Wali R, Tilley E et al. Replisome trafficking in growing vegetative hyphae of Streptomyces coelicolor A3(2). J Bacteriol. 2011;193:1273–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu LJ, Errington J. Coordination of cell division and chromosome segregation by a nucleoid occlusion protein in Bacillus subtilis. Cell. 2004;117:915–25. [DOI] [PubMed] [Google Scholar]
- Yang C-C, Huang C-H, Li C-Y et al. The terminal proteins of linear Streptomyces chromosomes and plasmids: a novel class of replication priming proteins. Mol Microbiol. 2002;43:297–305. [PubMed] [Google Scholar]
- Yang C-C, Sun W-C, Wang W-Y et al. Mutational analysis of the terminal protein Tpg of Streptomyces chromosomes: identification of the deoxynucleotidylation site. PLoS One. 2013;8:e56322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C-C, Tseng S-M, Pan H-Y et al. Telomere associated primase Tap repairs truncated telomeres of Streptomyces. Nucleic Acids Res. 2017;45:5838–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang MC, Losick R. Cytological evidence for association of the ends of the linear chromosome in Streptomyces coelicolor. J Bacteriol. 2001;183:5180–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y-H, Song E, Willemse J et al. A novel function of Streptomyces integration host factor (sIHF) in the control of antibiotic production and sporulation in Streptomyces coelicolor. Antonie Van Leeuwenhoek. 2012;101:479–92. [DOI] [PubMed] [Google Scholar]
- Yokoyama E, Doi K, Kimura M et al. Disruption of the hup gene encoding a histone-like protein HS1 and detection of HS12 of Streptomyces lividans. Res Microbiol. 2001;152:717–23. [DOI] [PubMed] [Google Scholar]
- Zhang L, Willemse J, Claessen D et al. SepG coordinates sporulation-specific cell division and nucleoid organization in Streptomyces coelicolor. Open Biol. 2016;6:150164. [DOI] [PMC free article] [PubMed] [Google Scholar]