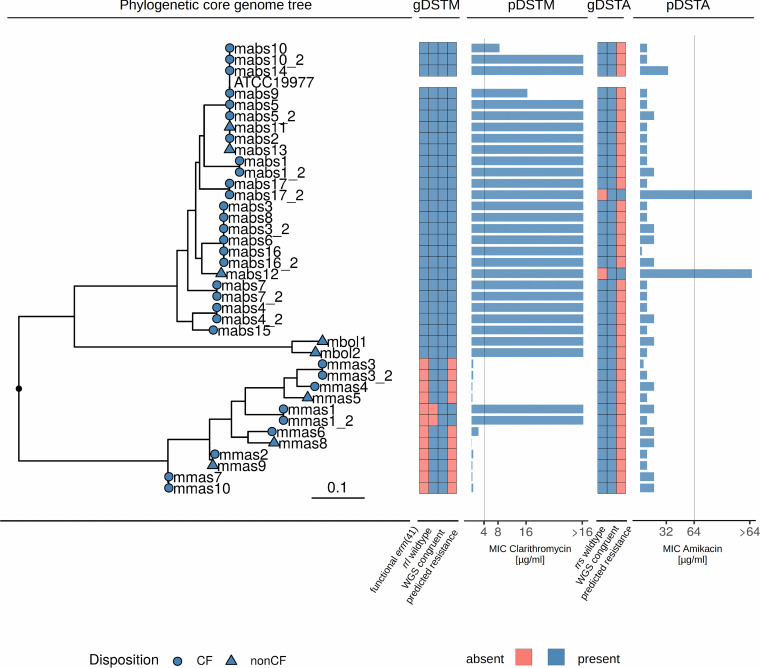
FIG 1.
Phylogenetic core genome tree with all isolates from our cohort (n = 39), heatmap of present or absent resistance genes for macrolide resistance (gDSTM), pDST results for clarithromycin (pDSTM), heatmap of gDST results for aminoglycosides (gDSTA), and pDST results for amikacin (pDSTA). Macrolide resistance was due mainly to functional erm(41) genes in M. abscessus subsp. abscessus. One rrl mutation could be found in an M. abscessus subsp. massiliense isolate. Two isolates showed mutations in the rrs gene and accordingly high MICs in amikacin. All predicted resistance levels by gDST (NTM-DR and WGS) were confirmed by pDST. Vertical lines in pDST panels depict breakpoints, as given by the CLSI. mabs, M. abscessus subsp. abscessus; mmas, M. abcsessus subsp. massiliense; mbol, M. abscessus subsp. bolletii. The tree scale depicts SNPs/site.