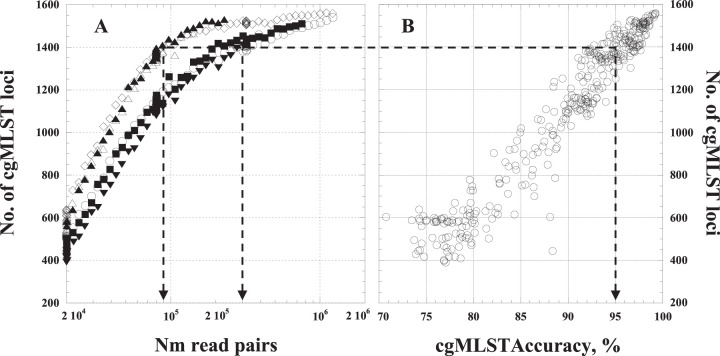
FIG 2.
Dependence of quality of N. meningitidis genome assemblies on the number of provided N. meningitidis read pairs. (A) The number of cgMLST loci present in genome assemblies as a function of the number of N. meningitidis read pairs used for the assembly. To produce a range of data set sizes, reads were randomly subsampled from the reads produced from the 6 enriched specimens for which genome assemblies had over 95% allele calling accuracy. Empty circles, Ur-2; empty diamonds, Ur-5; filled squares, Ur-7; filled triangles, Ur-9; empty triangles, Ur-10; filled inverse triangles, Ur-11. (B) Correlation between accuracy of allele calling for cgMLST loci and the number of loci present. The arrows shows correspondence between minimum tolerated allele calling accuracy (95%) and the minimum number of N. meningitidis read pairs or cgMLST loci required to provide that level of accuracy.