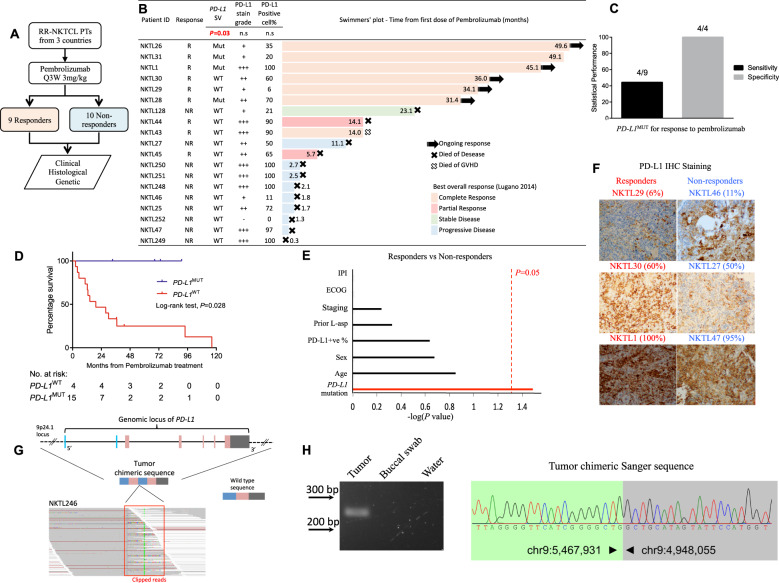
Fig. 1. PD-L1 structural rearrangements (PD-L1MUT) as a potential biomarker of response to pembrolizumab for patients with RR-NKTCL.
a Schematic makeup of the study and the stratification of patients with RR-NKTCL accordingly to their response to pembrolizumab. b Swimmers’ plot showing the duration of responses for the 19 patients with RR-NKTCL who were treated with pembrolizumab. Tabular data showing the PD-L1 mutation status, immunohistochemical (IHC) PD-L1 positivity of tumor cells and PD-L1 stain grade (− is negative, + is weakly stained, ++ is moderately stained and + ++ is strongly stained) accompanies each corresponding NKTCL sample. c Statistical performance measures of sensitivity and precision by PD-L1MUT as a predictor for responders to pembrolizumab. d Kaplan–Meier plot comparing the overall survival of patients with PD-L1MUT and PD-L1WT tumors. e Statistical tests on various clinical features and gene-mutation between responders and non-responders were carried out and the respective -log(P value) were plotted. The vertical red line denotes the cutoff for significance at P = 0.05. f Representative images of PD-L1 IHC weakly, moderately, strongly stained images for tumors from both responders and non-responders of patients with RR-NKTCL to pembrolizumab. Percentages of tumor cells positively stained by PD-L1 antibody are in brackets. g Schematic diagram of the wild-type 9p24.1 locus and the chimeric sequence representing the PD-L1MUT detected in the tumor DNA of NKTL246. A snapshot of the aligned sequencing reads, which are soft-clipped, at the genomic breakpoint of the PD-L1MUT are shown in the ‘red’ box. h PCR-based gel validation correctly amplified the 246 bp chimeric PD-L1 sequence from the tumor (T), and not from the buccal swab (BS), water (H20). Sanger sequence validated the chimeric PD-L1 to base-pair resolution. R responder, NR non-responder, MUT mutant, WT wild type, IPI international prognostic index, ECOG eastern cooperative oncology group, n.s. not significant.