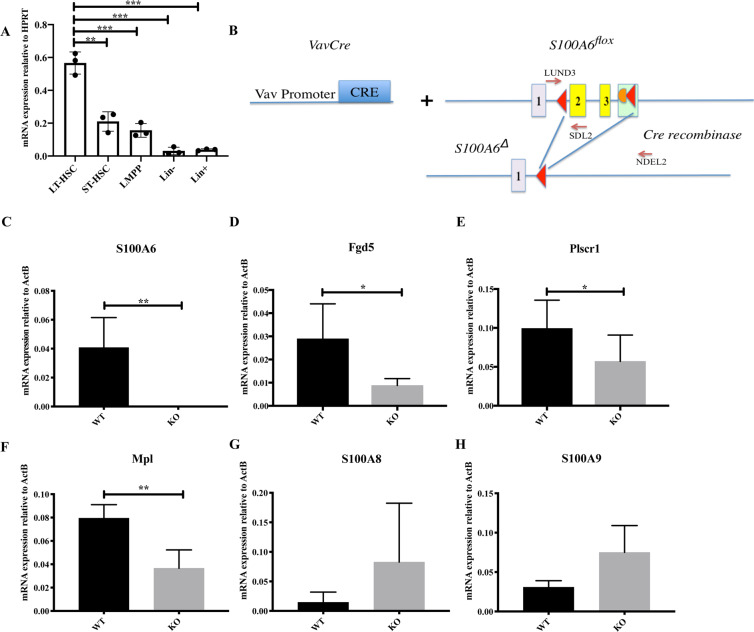
Fig. 1. S100A6 is highly expressed in LT-HSCs and several stem-cell-specific transcripts are decreased in the absence of S100A6.
a Quantitative real-time PCR (qRT-PCR) analysis of S100a6 expression in long-term HSC (LT-HSCs; (lineage−Scal-1+ c-Kit+) LSK CD34−Flt3−), short-term HSC (ST-HSC; LSK CD34+Flt3−), lymphoid-primed multipotent progenitors (LMPP; LSK CD34+Flt3+), lineage−, and lineage+ cells. Each value is normalized to HPRT expression and mean ± SD of triplicates is shown (n = 3; *p < 0.05; **p < 0.001; ***p < 0.0001; analyzed by an unpaired two-sided t-test). b Schematic representation of Vav-Cre-mediated conversion of the S100a6flox allele into the S100a6Δ allele by deletion of DNA between the two loxP sites in the S100a6flox locus. This includes the entire S100a6 exons 2 and 3 (yellow). Exons 1–3 are indicated. Red triangles are loxP sites. Orange semisphere is FRT site from excised neo cassette (green rectangular). Cre recombinase cleaved at loxP sites. c S100a6 mRNA expression in BM HSCs (CD150+, CD48−, Flt3−, CD34−) (n = 4). mRNA levels of Fgd5 (n = 4) (d), Plscr1 (n = 8) (e), Mpl (n = 4) (f), S100A8 (n = 4) (g), S100A9 (n = 3) (h), assessed by qRT-PCR on LT-HSCs (CD150+CD48−CD34−Flt−). Results are the mean ± SD of triplicates. Each value is normalized to ActB expression (*p < 0.05; **p < 0.001; analyzed by an unpaired two-sided t-test).