Abstract
Stress granules (SGs) are non-membranous cytosolic protein-RNA aggregates that process mRNAs through stalled translation initiation in response to cellular stressors and in disease. DEAD-Box RNA helicase 3 (DDX3) is an active target of drug development for the treatment of viral infections, cancers, and neurodegenerative diseases. DDX3 plays a critical role in RNA metabolism, including SGs, but the role of DDX3 enzymatic activity in SG dynamics is not well understood. Here, we address this question by determining the effects of DDX3 inhibition on the dynamics of SG assembly and disassembly. We use two small molecule inhibitors of DDX3, RK33 and 16D, with distinct inhibitory mechanisms that target DDX3’s ATPase activity and RNA helicase site, respectively. We find that both DDX3 inhibitors reduce the assembly of SGs, with a more pronounced reduction from RK-33. In contrast, both compounds only marginally affect the disassembly of SGs. RNA-mediated knockdown of DDX3 caused a similar reduction in SG assembly and minimal effect on SG disassembly. Collectively, these results reveal that the enzymatic activity of DDX3 is required for the assembly of SGs and pharmacological inhibition of DDX3 could be relevant for the treatment of SG-dependent pathologies.
Keywords: DDX3, DDX3X, Stress Granule, RK-33, DEAD-Box RNA helicase
1. Introduction
Translational arrest is a common stress response in eukaryotic cells that results in the cytosolic accumulation of non-membranous, untranslated ribonucleoprotein (RNP) aggregates called stress granules (SGs) [1–3]. Their assembly is triggered by various intracellular [4] and extracellular stressors such as reactive oxygen species, heat, starvation, and viral infection [5]. SGs are highly dynamic structures, which rapidly assemble and disassemble in coordination with translational arrest [6, 7]. Alterations in SG dynamics are linked to various pathological conditions. For example, stable SGs are believed to be associated with the pathogenesis of neurodegenerative diseases and cancers [2, 8–10]. SGs also play a dual role in viral infection: they are involved in innate immune response, but can also be hijacked by the viruses to switch translational machinery to the production of viral proteins [11]. As such, SGs have been suggested as a target for treatments of these wide-ranging diseases [12, 13].
SGs consist of stalled translation preinitiation complexes such as small 40S ribosomal subunits, translation initiation factors, and proteins with intrinsically disordered regions (IDRs) [14, 15]. One of the most well-studied and critical proteins for SG assembly is Ras GTP-activating protein (G3BP1) [13, 16–19]. The members of the DEAD-Box RNA helicase family, eIF4A and DEAD-Box RNA helicase 3 (DDX3) [7], are also defined as markers and core components of SGs.
Recent evidence suggests that DDX3 might be a major regulatory component of SGs [20]. Moreover, disruption of DDX3 functions has been found to be associated with the formation of stable, pathological SGs [21]. In mammals, DDX3 is encoded by two highly homologous genes: X chromosomal DDX3X and Y chromosomal DDX3Y. This dsRNA helicase consists of two domains connected by a flexible linker, and both domains contribute to DDX3 ATPase and RNA helicase enzymatic activities [22, 23]. While inhibition of helicase activity does not affect ATPase activity of the enzyme, inhibition of ATPase activity by chemical inhibitors or point mutations completely abolishes helicase activity [24, 25].
Because DDX3 is involved in gene-specific transcriptional and translational regulation [26–30], dysregulation of DDX3 activity is not surprisingly associated with many pathological processes [31]. For example, DDX3 has been associated with the propagation of human viruses including HIV [32, 33], and tumor development and progression [34]. Therefore, DDX3 has been proposed as a target for anti-cancer and anti-viral therapy, and several small molecule inhibitors have been developed and tested in animal studies. Two of the most advanced inhibitors are RK-33 [25] and 16D [33]. RK-33 is a potent small-molecule inhibitor of the ATP binding cleft of DDX3 [25]. RK-33 has been shown to strongly inhibit the ATPase activity of DDX3, suppress tumor growth, and sensitize cancer to radiation therapy. 16D has been shown to competitively occupy the RNA binding site of DDX3, suppressing the RNA helicase activity of DDX3 without affecting its ATPase activity, and 16D exhibits broad-spectrum antiviral activity in vitro [33]. Both compounds were safely tested in animal models and did not induce abnormal clinical chemistry markers or histological profiles [25, 33]. Previously we showed that RK-33 protects neurons from combined HIV-Tat and cocaine-induced toxicity, and acts through inhibiting microglia activation. This result suggests that DDX3 inhibition should be further explored as a treatment of HIV-associated neurocognitive disorder, along with other neurodegenerative diseases [31].
Here, we address the effects DDX3 enzymatic activities have on the dynamics of SGs by using two different pharmacological inhibitors and siRNA-based depletion of DDX3. Our results show that inhibition of DDX3 enzymatic activity and knockdown of DDX3 decrease the rate of SG assembly without strongly affecting the SG disassembly. Longer term, our study points to pharmacological inhibition of DDX3 as a strategy for treatment of SGs-related diseases.
2. Materials and Methods
2.1. Cell culture
U2OS human osteosarcoma cells (American Type Culture Collection) were grown in high glucose Dulbecco’s modified Eagle’s medium (American Type Culture Collection, Manassas, VA) supplemented with 10% fetal bovine serum (Mediatech Inc., Manassas, VA) and 1% penicillin/streptomycin solution (HyClone reagents from GE Healthcare Life Sciences, Pittsburgh, PA). Cells were cultured at 37°C in a humidified atmosphere with 5% CO2. During cell passaging, cells were checked for the presence of mycoplasma contamination using MycoFluor Mycoplasma detection kit (Thermo Fisher Scientific, Waltham, MA). Cell line authentication was performed at the University of Arizona Genetics Core (Tucson, AZ).
2.2. Drugs and chemical reagents
RK-33 (Cas No. 1070773–09-9) was purchased from Selleck Chemicals (S8246, Houston, Texas). A stock solution of RK-33 was prepared in 5 mM DMSO (Millipore Sigma, D8418) and was diluted to final concentration of 6 μM. 16D (CAS No. 1919828–83-3) was kindly provided by Drs. Giovanni Maga and Maurizio Botta (Institute of Molecular Genetics IGM-CNR “Luigi Luca Cavalli-Sforza”, National Research Council, via Abbiategrasso 207, I-27100 Pavia (Italy)). A 1 mM stock solution of 16D was prepared in DMSO and was diluted to the final concentration of 48 µM. Sodium arsenite, referred to as arsenite for simplicity, was purchased from Millipore Signa (S7400). A 0.05 M stock solution of arsenite was prepared in water and was diluted to final concentration of 0.5 mM.
2.3. DDX3X plasmid construct
The wild-type DDX3X construct tagged with FLAG was donated by David Sabatini (Addgene plasmid# 70648; http://n2t.net/addgene: 70648; RRID:Addgene_70648). The plasmid was isolated and collected using ZymoPURE Plasmid Miniprep Kit (Zymo Research Cat.# 4210). Mutations were established using Q5® Site-Directed Mutagenesis Kit Protocol (New England Biolabs Cat.# E0554). Site-directed mutation was established via whole-plasmid amplification.
The primer sequences for wild-type DDX3X active sites were:
5’-TCGAACAAGATACTATGCCTCCAAAGGGTGTCCGC-3’
5’-CTATTCTACGAATCTGAGGCTCAAACCCCATATCCAACATCCG-3’
The primer sequences to establish mutant DDX3X active sites were:
5’-TCGAACAAGATACTATGCCTCCAAAGGGTGTCCGCCACACTATGATGTTTGCTGCT GCTTTTCCTAAGGA-3’
5’-CTATTCTACGAATCTGAGGCTCAAACCCCATATCCAACATCCGATCAGCTTGATCTA ACACCAA-3’
The primer sequences to establish knockout DDX3X active sites were:
5’-TATGAACACCACTACAAGGG-3’
5’-AGCATGCTTTTGCACTGGAG-3’
Ligation to reestablish a circular plasmid after PCR was performed with kinase, ligase, and Dpnl (KLD) enzyme mix and reaction buffer also found in the Q5® Site-Directed Mutagenesis Kit from New England Biolabs. Successful mutation of the plasmid was verified by Sanger sequencing.
2.4. Experimental designs
For SG assembly experiments (Fig. 1, 3), cells were either vehicle treated or pre-treated with 6 µM RK-33 or 48µM 16D for 1 h. Cells were then treated with 0.5 mM arsenite in the 37 degree C incubator for 30 min, 60 min, and 90 min. For siRNA SG assembly experiments (Fig. 5), cells were directly treated with 0.5 mM arsenite for 30 min, 60 min, and 90 min. For SG disassembly experiments (Fig. 2, 3), cells were pre-treated with 0.5 mM arsenite for 1 h, then the arsenite-containing media was replaced with DMEM/10% FBS/1% pen-strep media lacking or containing 6 µM RK-33 for 30 min, 60 min, and 90 min. For siRNA SG disassembly experiments (Fig. 5), cells were treated with 0.5 mM arsenite for 1 h, then the arsenite-containing media was replaced with plain media as previously described.
Fig. 1.
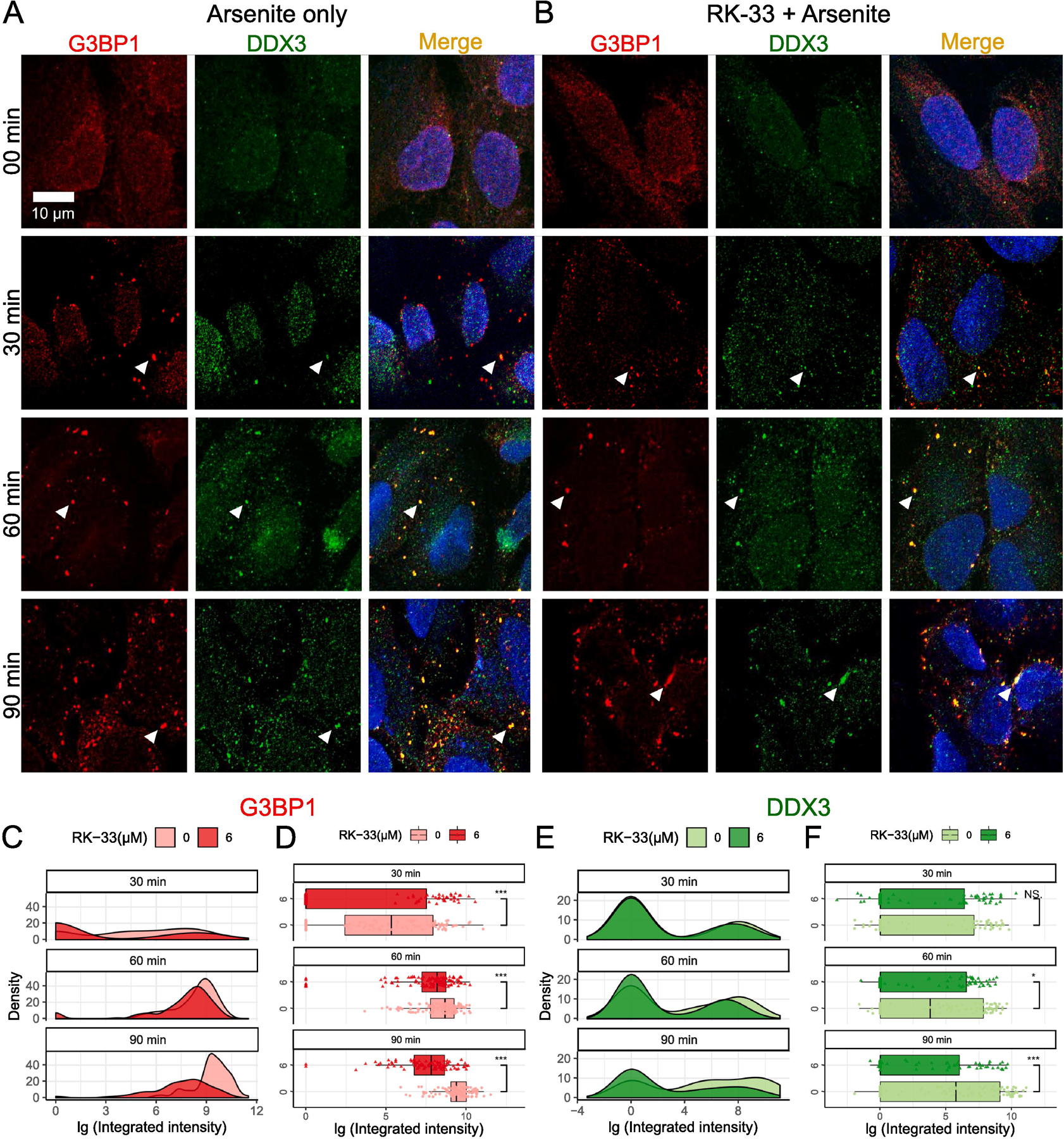
Effects of RK-33 treatment on SG assembly. (A–B) SG assembly was examined by Immunofluorescence with anti-G3BP1 (red) and anti-DDX3 (green). Scale bar, 10 µm. Cells were subjected to 00 min, 30 min, 60 min, and 90 min of 0.5 mM arsenite stress either in the (A) absence or (B) presence of 6 µM RK-33 pre-treatment for 1 h. The results of the integrated intensity are presented in log10 scale units, and graphically visualized in the form of density plots on the right and box-and-whisker plots with corresponding statistical parameters on the left. (C and D) Time course distribution of G3BP1 granules either in absence (lighter red) or presence (darker red) of RK-33 pre-treatment. (E and F) Time course distribution of DDX3 granules either in absence (lighter green) or presence (darker green) of RK-33 pre-treatment. (C and E) shows density plots and (D and F) shows box-and-whisker plots. The boxes cover 50% of data, and lines within boxes indicate median values. The Wilcoxon Signed-Rank Test was used to calculate statistical significance (* p < 0.05; ** p < 0.01; *** p < 0.001). The arrowheads represent endogenous G3BP1-dependent SGs.
Fig. 3.
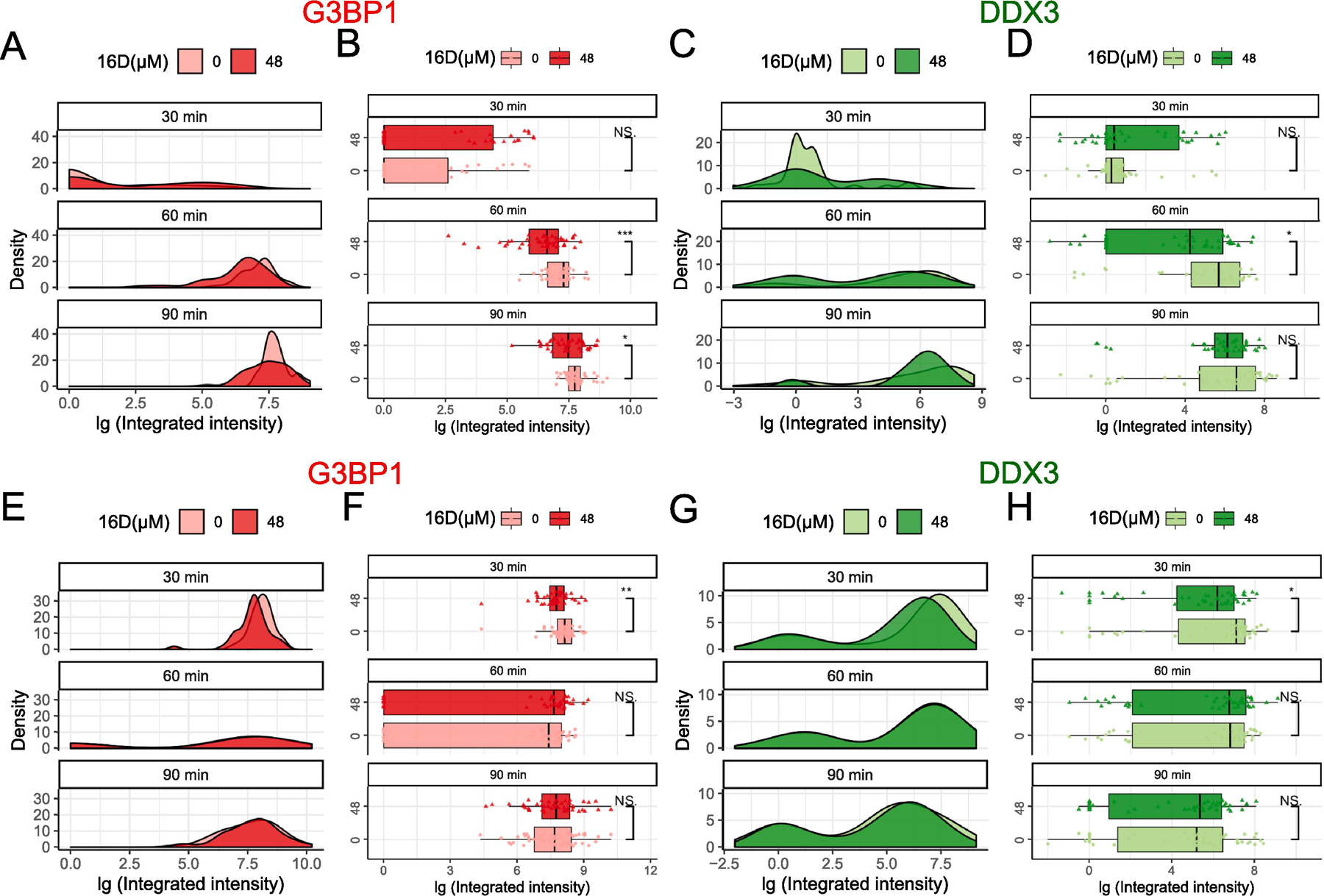
Effects of 16D on SG assembly and disassembly. (A–D) 16D on SG assembly was examined by subjecting U2OS cells to 30 min, 60 min, and 90 min of 0.5 mM arsenite stress either in the absence or presence of 48 µM 16D pre-treatment for 1 h. (E–H) 16D on SG disassembly was examined subjecting cells to 1 h of 0.5 mM arsenite pre-treatment followed by media replacement to recover for 30 min, 60 min, and 90 min in absence or presence of 48 µM 16D. (A–B, E–F) Time course distribution of G3BP1 granules either in absence (lighter red) or presence (darker red) of 16D treatment. (C–D, G–H) Time course distributions of DDX3 granules either in absence (lighter green) or presence (darker green) of 16D treatment. The boxes cover 50% of data, and lines within boxes indicate median values. The Wilcoxon Signed-Rank Test was used to calculate statistical significance (* p < 0.05; ** p < 0.01; *** p < 0.001).
Fig. 5.
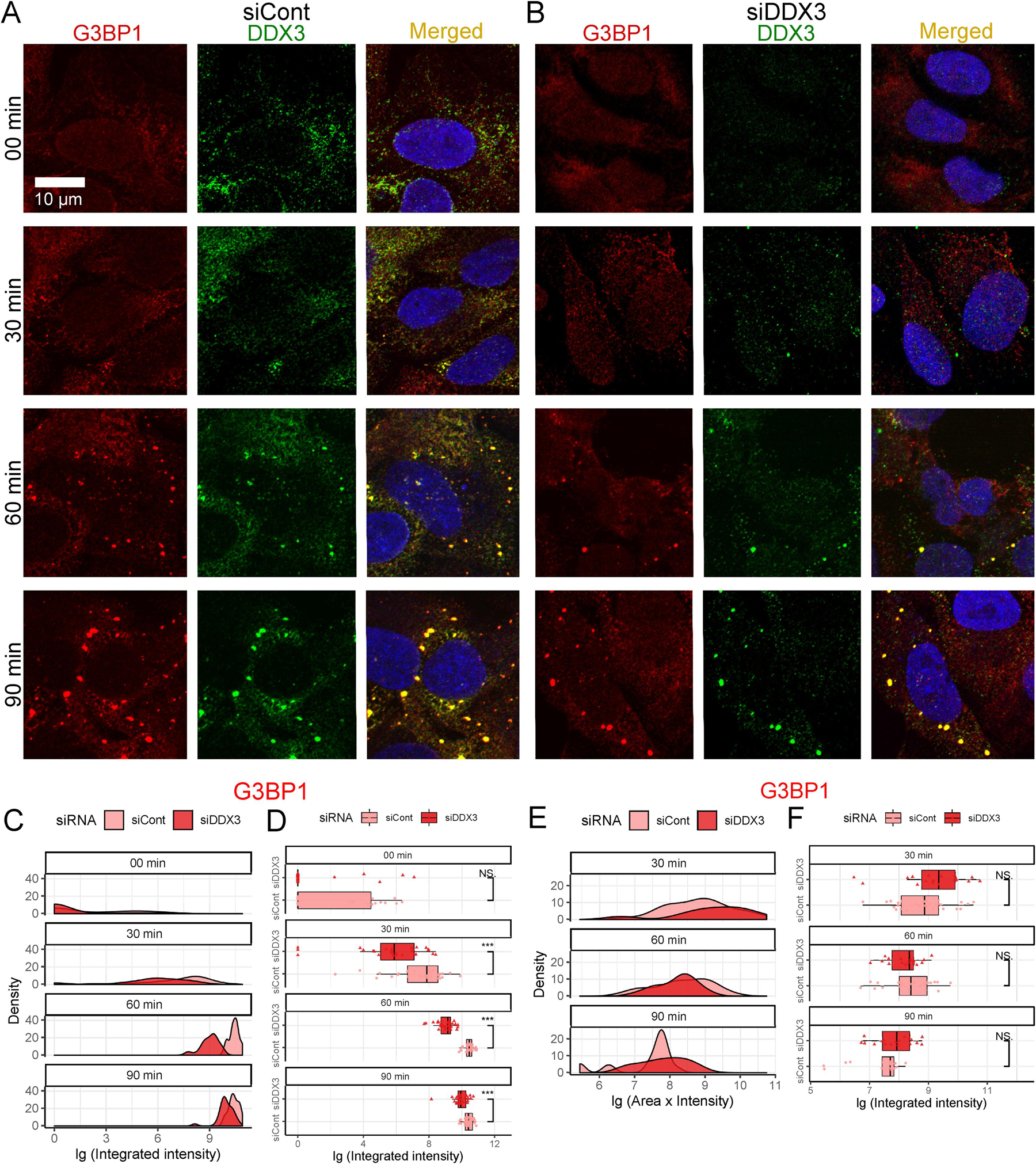
Effects of DDX3 siRNA knockdown on SG assembly and disassembly. (A–B) SG assembly was examined by Immunofluorescence with anti-G3BP1 (red) and anti-DDX3 (green). Scale bar, 10 µm. Cells subjected to 00 min, 30 min, 60 min, and 90 min of 0.5 mM arsenite stress either (A) without or (B) with DDX3 siRNA knockdown are quantified in (C and D). Cells were subjected to 60 min of 0.5 mM arsenite treatment followed by replaceing with fresh media to recover for 30 min, 60 min, and 90 min without or with DDX3 siRNA knockdown and are quantified in (E and F). (C–F) Time course distributions of G3BP1 granules either without (lighter red) or with (darker red) DDX3 siRNA knockdown. (C and E) shows density plots and (D and F) shows box-and-whisker plots. The boxes cover 50% of data, and lines within boxes indicate median values. The Wilcoxon Signed-Rank Test was used to calculate statistical significance (* p < 0.05; ** p < 0.01; *** p < 0.001).
Fig. 2.
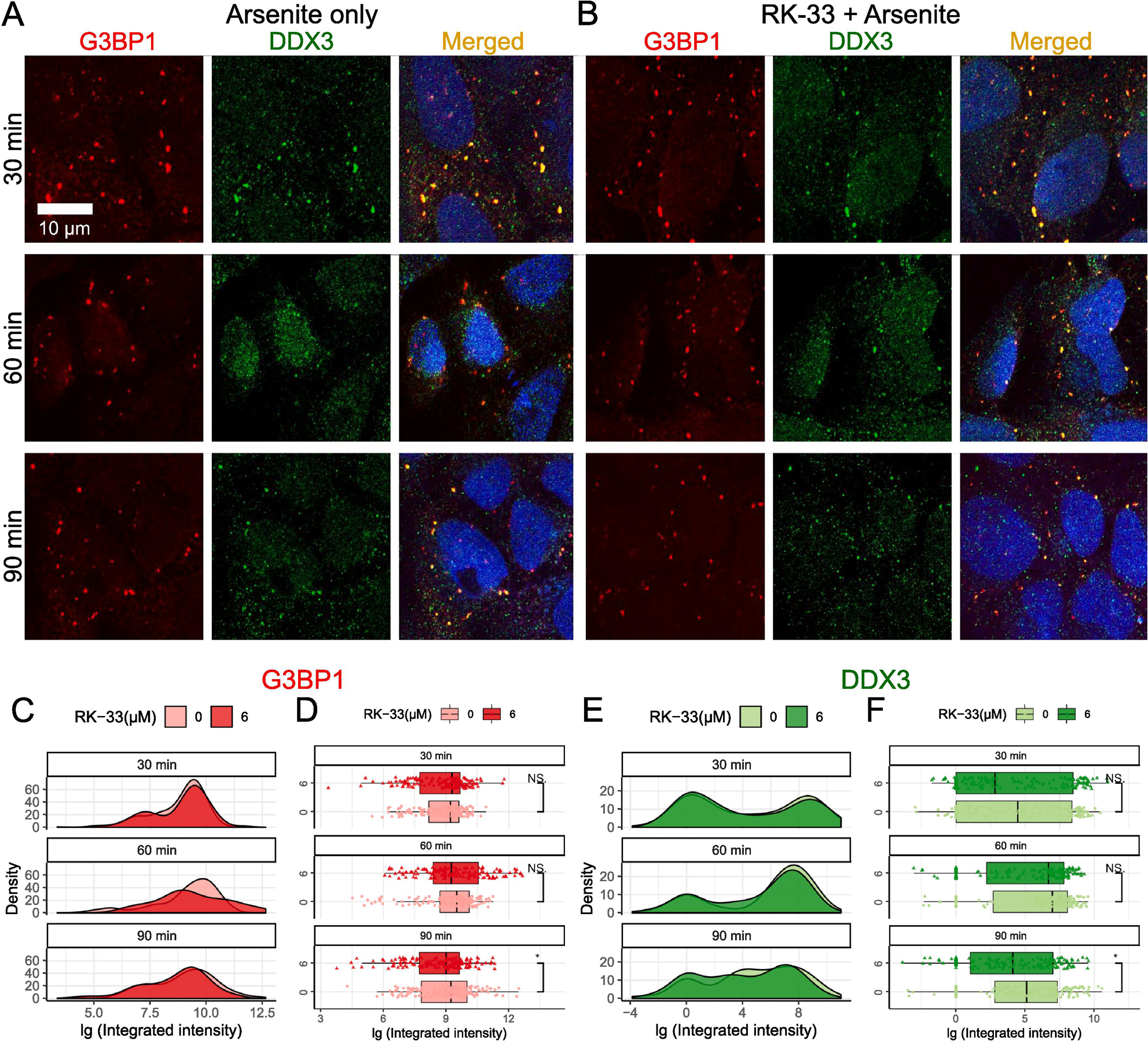
Effects of RK-33 treatment on SG disassembly. (A–B) SG disassembly was examined by Immunofluorescence with anti-G3BP1 (red) and anti-DDX3 (green). Scale bar, 10 µm. Cells were subjected to 1 h of 0.5 mM arsenite pre-treatment followed by media replacement to recover for 30 min, 60 min, and 90 min in the (A) absence or (B) presence of 6 µM RK-33. (C and D) Time course distributions of G3BP1 granules either in absence (lighter red) or presence (darker red) of RK-33. (E and F) Time course distributions of DDX3 granules either in absence (lighter green) or presence (darker green) of RK-33. (C and E) shows density plots and (D and F) shows box-and-whisker plots. The boxes cover 50% of data, and lines within boxes indicate median values. The Wilcoxon Signed-Rank Test was used to calculate statistical significance (* p < 0.05; ** p < 0.01; *** p < 0.001).
2.5. Image acquisition, processing, and analysis
Images were taken using a Carl Zeiss LSM 700 laser scanning confocal microscope controlled by ZEN Black software (Carl Zeiss Meditec, Dublin, CA) and equipped with 20x (Plan APO 0.8 air) or 63x (Plan APO 1.4 oil DIC) objectives unless otherwise stated. Fluorescence was excited using 405 nm, 488 nm, and 555 nm lasers. Single 16-bit depth images were captured using near 1 AU pinhole size and 1.0 scanning zoom with 312-nm (20x) or 142-nm (63x) X-Y pixel size. Laser power and gain settings were selected to maximize a signal-to-noise ratio within each individual fluorescence channel and cover the potential dynamic range of the correspondeing fluorescence signal. Fluorescence and differential interference contrast (DIC) imaging was done using single-frame or tilling modes. ImageJ software (National Institutes of Health, USA) was used for manual or automatic analysis of microscopy images acquired using a Zeiss 700 confocal microscope. The total number of G3BP1 and DDX3 granules were estimated using automatic segmentation of the corresponding fluorescence channels. Both channels were subjected to bandpass filtration, background subtraction, thresholding, and particle analysis using lower (4 pixels) cutoff size. The sequence of operations and the parameter values used were verified using randomly selected subsets of images. The G3BP1 and DDX3 intensity thresholds were selected manually to ensure detection of bright SG and minimization of random noise contribution. The number of cells within the field of view was estimated using automatic segmentation of a denoised DNA channel (DAPI) followed by “Analyze Particles” ImageJ command. High frequency noise was attenuated by a low-pass filter (“Gaussian Blur” with a parameter sigma set to 3). The cutoff size (1000 pixels) was selected by visual examination of the microscopy images to exclude from consideration cell debris. Data were aggregated, analyzed, and visualized using MatLab (MathWorks) and R ggplot2 tools.
Numbers of G3BP1- and DDX3-positive SGs were defined by direct counts obtained using the “Analyze Particles” ImageJ tool. Amounts of G3BP1 and DDX3 proteins accumulated within a single granule were estimated by a total intensity of the corresponding fluorescence channel (Area of SG x Mean Intensity of SG). Here, we assumed that the fluorescence intensity of an SG is proportional to the amount of protein accumulated, therefore, we refer to this area multiplied by fluorescence intensity as “integrated intensity” throughout the paper. These values were used as a convenient marker of SG assembly and disassembly. The corresponding distributions and statistical measures were obtained using R programming language
2.6. Immunofluorescence staining
The Immunofluorescence staining of U2OS cells was performed as previously described [31]. Briefly, cells were plated on glass coverslips that were placed inside 12-well plates. After the experimental treatments, cells were fixed with 4% paraformaldehyde and permeabilized with 0.1% Triton X-100. The fixed cultures were blocked with 10% fetal bovine serum for 1 h and then co-labeled overnight with different primary antibodies: rabbit polyclonal anti-DDX3 (1:300) (A300–474A, Bethyl, Montgomery, TX), mouse monoclonal anti-DDX3 (1:300) (sc-365768, Santa Cruz Biotechnology, Dallas, TX), rabbit polyclonal anti-G3BP1 (1:500) (A302–033A, Bethyl, Montgomery, TX), or mouse monoclonal anti-FLAG M2 (1:2000) (F1804, Sigma-Aldrich, St. Louis, MO). The fixed cultures were then incubated for 1 h with following secondary antibodies all from Thermo Fisher Scientific: donkey anti-rabbit IgG conjugated with AlexaFluor 488 (1:500) (A-21206), goat anti-mouse IgG conjugated with AlexaFluor 594 (1:500) (A-11005), donkey anti-rabbit IgG conjugated with AlexaFluor 594 (1:500) (A-21207), or donkey anti-mouse IgG conjugated with AlexaFluor 488 (1:500) (A-21202). To identify cell nuclei, DAPI was added with the final PBS wash, and coverslips were mounted on glass slides using VECTASHIELD Vibrance mounting medium (Vector Laboratories, Burlingame, CA).
2.7. siRNA transfection
siRNA transfection was performed using a reverse transfection method as previously [35]. According to the manufacture specifications, Opti-MEM media (ThermoFisher Scientific, Waltham, MA) was premixed with siLentFect (Bio-Rad, Hercules, CA). siRNAs were added at the protocol recommended final concentration and the siLentFect/Opti-MEM/siRNA mixture was incubated at room temperature for 20–30 min. The premixed siRNA/siLentFect complexes were added to cells at a seeding density of 106 cells per well in 6 well plate. siRNA treated cells were grown for 3–4 days at 37 °C and 5% CO2.
2.8. Statistics and data analysis
Statistical analyses were carried out using R version 3.6.1 and RStudio version 1.2.5001.
Experiments were performed with independent triplicates.
The density plot was created using ggplot2 package, in particular, the geom_density() function.
The box-and-wisker plot was created using the geom_boxplot() function from the ggplot2 package and the geom_signif() function from the ggsignif package. The boxes cover 50% of data, and lines within boxes indicate median values.
Normally distributed samples were calculated using parametric analysis method. If there was a deviation from a normal distribution, non-parametric analysis methods were used. Statistical differences between two groups in each time group were evaluated by Wilcoxon Signed-Rank Test using geom_signif(test=“wilcox.text”). P < 0.05 was considered significant. All relevant data were expressed as mean ± SEM.
Statistical plots and figures were generated using Inkscape version 0.92 and the following R packages: dplyr version 0.8.3 (for initial data organization and individual statistical tests), data.table version 1.12.2 (for eventual data organization and dataframe construction), ggplot2 version 3.2.1 (for data visualization), ggsignif version 0.6.0 (for eventual statistical evaluations).
2.9. Western blot
Cells were lysed using RIPA lysis buffer (50 mM Tris-HCl, pH 7.4; 150 mM NaCL; 5 mM EDTA; 0.5 mM EGTA; 1% Igepal CA-630; 0.5% sodium deoxycholate; and 0.1% SDS) and sonicated. Protein concentration was measured using Bio-Rad DC protein assay (Bio-Rad). 20 µg of proteins were separated in precast 4–12% polyacrylamide gels (M00654, GeneScript) and transferred onto PVDF membranes (88518, Thermo Fisher Scientific). The membranes were then stained with Ponceau Red, blocked for 1 h in 1X TBS-T buffer (20 mM Tris-HCl, pH 7.6; 140 mM NaCl; 0.1% Tween-20) with 5% non-fat dry milk, and incubated with primary antibodies overnight at 4 °C. The primary antibodies were rabbit polyclonal anti-DDX3 (1:3000) (A300–474A, Bethyl, Montgomery, TX), mouse monoclonal anti-DDX3 (1:3000) (sc-365768, Santa Cruz Biotechnology, Dallas, Texas), rabbit monoclonal anti-B-actin (1:10,000) (D6A8, Cell Signaling Technology, Danvers, MA), and rabbit monoclonal anti-GAPDH (1:10,000) (14C10, Cell Signaling Technology, Danvers, MA). The secondary antibodies were HRP-conjugated anti-mouse IgG (1:8000) (7076S) and HRP-conjugated anti-rabbit IgG (1:8000) (7074S), both from Cell Signaling. Membranes were processed with SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Scientific).
3. Results
3.1. RK-33 significantly modulates SG assembly
To evaluate the importance of DDX3 enzymatic activity on SG assembly, we used RK-33, an inhibitor of DDX3 ATPase activity, and quantified the dynamics of canonical G3BP1-positive SGs by employing a well-established SG induction model: the induction of oxidative stress in U2OS osteosarcoma human cell line using 0.5 mM sodium arsenite, hereafter referred to as arsenite for simplicity, treatment [9, 17, 36]. U2OS cells are well-established as a cell model to explore subcellular localization of SGs [16, 37]. U2OS cells are widely used because their large and flat morphology facilitate immunofluorescence studies of SGs. Additionally, SG dynamics observed in U2OS cells have been entirely consistent with SG dynamics extensively investigated in many other cell lines [38]. Accordingly, to determine the effect of RK-33 on SG assembly, U2OS cells were pre-treated with 6 µM RK-33 or vehicle control for 1 h followed by arsenite treatment and the resulting SGs were visualized using G3BP1 or DDX3 antibodies at three different stress time points (Fig. 1A–B). The measurement of G3BP1 localization was chosen because of its established connection with SG dynamics [13, 17, 19, 39]. The concentration of RK-33, 6 µM, was selected based on our previous and other relevant studies [25, 31].
Fig. 1A–B illustrate formation of SGs during the first 90 min of arsenite treatment. Pre-treatment of 1 h RK-33 did not cause any distinct accumulations of GRBP1 (red) and DDX3 (green) (Fig. 1A–B, 00 min). The graphical distribution of SG assembly for the starting time point (00 min) is not shown as there were no detectable SGs in the absence of arsenite. After 30 min of arsenite stress, however, cells began to demonstrate bright G3BP1 and DDX3 granules characteristic of SGs (arrowheads, Fig. 1A–B, 30 min). Prolonged arsenite exposures led to a significant increase in the number, size, and brightness of SGs (Fig. 1A–B, 60–90 min). We quantified these condensates using total G3BP1 or DDX3 fluorescence signals of individual granules; we refer to this value as “SG integrated intensity” hereafter (detailed in Materials and Methods) (Fig. 1C–F) [40, 41]. The integrated-intensity values are presented in log10 scale units, indicated as “lg”, and graphically visualized in the form of density plots on the right (Fig. 1C and E) and box-and-whisker plots with corresponding statistical parameters on the left (Fig. 1D and F).
G3BP1-positive granules induced by arsenite were markedly reduced in U2OS cells that were pre-treated with 6 µM RK-33 1 h (Fig. 1A–B). G3BP1 density plots of SG integrated intensities showed peaks in RK-33 pre-treatment groups, represented by the dark red shade, all shifted leftward compared to those of vehicle control groups, represented by the light red shade (Fig. 1C). The difference between the drug pre-treated and vehicle pre-treated groups was particularly noticeable by 90 min, in which the log10 of SG integrated intensity for RK-33 pre-treated group predominantly fell within the 0–9 range, whereas the SG integrated intensity of the vehicle pre-treated group fell within the 9–12 range (Fig. 1A–B, 90 min). Consequently, the median integrated intensity shown in 90 min box plots for the RK-33 pre-treatment group was significantly lower than that of the control group, and the same statistical significance was observed for all other time points (p<0.001) (Fig. 1D).
DDX3 granules at 60 min and 90 min showed significant reductions of SG integrated intensities in the RK-33 pre-treatment group compared to those of the control (Fig. 1F). Interestingly, DDX3-positive granules showed distinct populations of small and large particles (i.e., bimodality) (Fig. 1E). Small granules were enriched at 30 min data point, and larger granules began to accumulate at 60 min and became prevalent by 90 min (Fig. 1E). Taken together, our comparison of the SGs assembled between RK-33 pre-treated and vehicle control pre-treated groups reveal that pharmacological inhibition of DDX3’s catalytic ATPase site suppresses canonical G3BP1-SG assembly.
3.2. RK-33 marginally modulate SG disassembly
The results above prompted investigation for the pharmacological intervention of DDX3 on SG disassembly, therefore, the effect of RK-33 on the rate of G3BP1-dependent SG disassembly using arsenite pre-treatment was evaluated. In a normal SG process, removal of the stressor leads to a disassembly of the SGs. Therefore, after 1 h of arsenite pre-treatment, we replaced arsenite-containing media with media containing 6 µM RK-33 or vehicle control and left to recover for 30 min, 60 min, and 90 min. The resulting G3BP1 immunofluorescence images did not show a noticeable difference between RK-33 treated and vehicle control treated cells (Fig. 2A–B). The G3BP1 density plots showed a general overlap between these two experimental groups (Fig. 2C). The boxplots of G3BP1 granules did not show any statistically significant difference for the groups treated with RK-33 at 30 min and 60 min time points, whereas a minor difference was noted in the 90 min group (Fig. 2D). More specifically, the median integrated intensity was significantly lower in the RK-33 treated group relative to the control in 90 min group (p<0.05) (Fig. 2D, 90 min). DDX3 granules continued to display their bimodal distribution in (Fig. 2E) and showed similar granule dissociation trends in the presence of RK-33 similar to those of the G3BP1 granules, with 90 min showing statistical significance (Fig. 2D and F). These results indicate that RK-33 suppresses the assembly of SGs, but only marginally affects disassembly.
3.3. 16D moderately modulates SG assembly and marginally modulates SG disassembly
The inhibition of the ATPase activity of DDX3 affects not only coupled dsRNA helicase activity, but also interactions with dsRNAs and other proteins [24, 42]. Here, we explored the effects of inhibition of DDX3’s helicase activity on SG assembly and disassembly. We tested 16D, a DDX3 inhibitor that specifically inhibits the DDX3 helicase enzymatic activity but not its ATPase activity [33], on SG dynamics utilizing the same conditions as above. Accordingly, U2OS cells were pre-treated with 1 h 48 µM 16D or vehicle control, followed by the treatment to 0.5 mM arsenite for 30 min, 60 min, and 90 min.
Although not as striking, the effects of 16D on SG assembly echoed that of RK-33. After 30 min of arsenite, no significant difference in SG assembly between the cultures lacking or containing 16D were detected (Fig. 3A–B). After 60 min, the median integrated intensity of 16D group was significantly less than that of the control group (Fig. 3B, 60 min). A similar trend was observed after 90 min (Fig. 3B, 90 min). Here too, we observed a bimodal distribution of DDX3 in the density plot (Fig. 3C), which was distinct from the single peak dominant G3BP1 (Fig. 3A). The statistical significance of the difference between the distribution of DDX3 in the absence or presence of 16D was not as evident, with only 60 min arsenite treatment samples deemed statistically significant with p<0.05 (Fig. 3C–D). As such, inhibition of DDX3 helicase activity with 16D provides a similar but less pronounced effect on SG assembly in comparison with RK-33 inhibition of ATPase activity and DDX3 protein depletion by siRNA.
Consistent with the results from RK-33 inhibitor, 16D treated cells did not show an increased rate for SG disassembly. The cultures pre-treated with arsenite and left to recover in an increasing time duration in the absence or presence of 16D showed minor differences for 30 min of recovery but not for 60 min and 90 min in both G3BP1 and DDX3 granule distributions (Fig. 3E–H). Therefore, targeting the RNA helicase activity with 16D dampens the formation of SG, with marginal effects on SG disassembly.
3.4. DDX3 siRNA knockdown significantly affected SG assembly and not SG disassembly
To further confirm the role of DDX3 in SG assembly, we analyzed the effect of the DDX3 knockdown on the dynamics of SGs in U2OS cells. Because the U2OS cell line is a female derived osteosarcoma cell line, it expresses only the X-chromosome encoded DDX3X isoform. To biochemically recapitulate the effect of DDX3 functional attenuation, DDX3X specific siRNAs, labeled as siDDX3, were compared with the effects of the scrambled siRNA control, labeled as siCont. The siRNA-specific downregulation of DDX3 expression was verified by western blot analysis (Fig. 4). To determine the effects of DDX3 depletion on SG formation, 48 h post-transfected U2OS cells were treated with 0.5 mM of arsenite as above, and the efficiency of SG assembly was quantified.
Fig. 4.
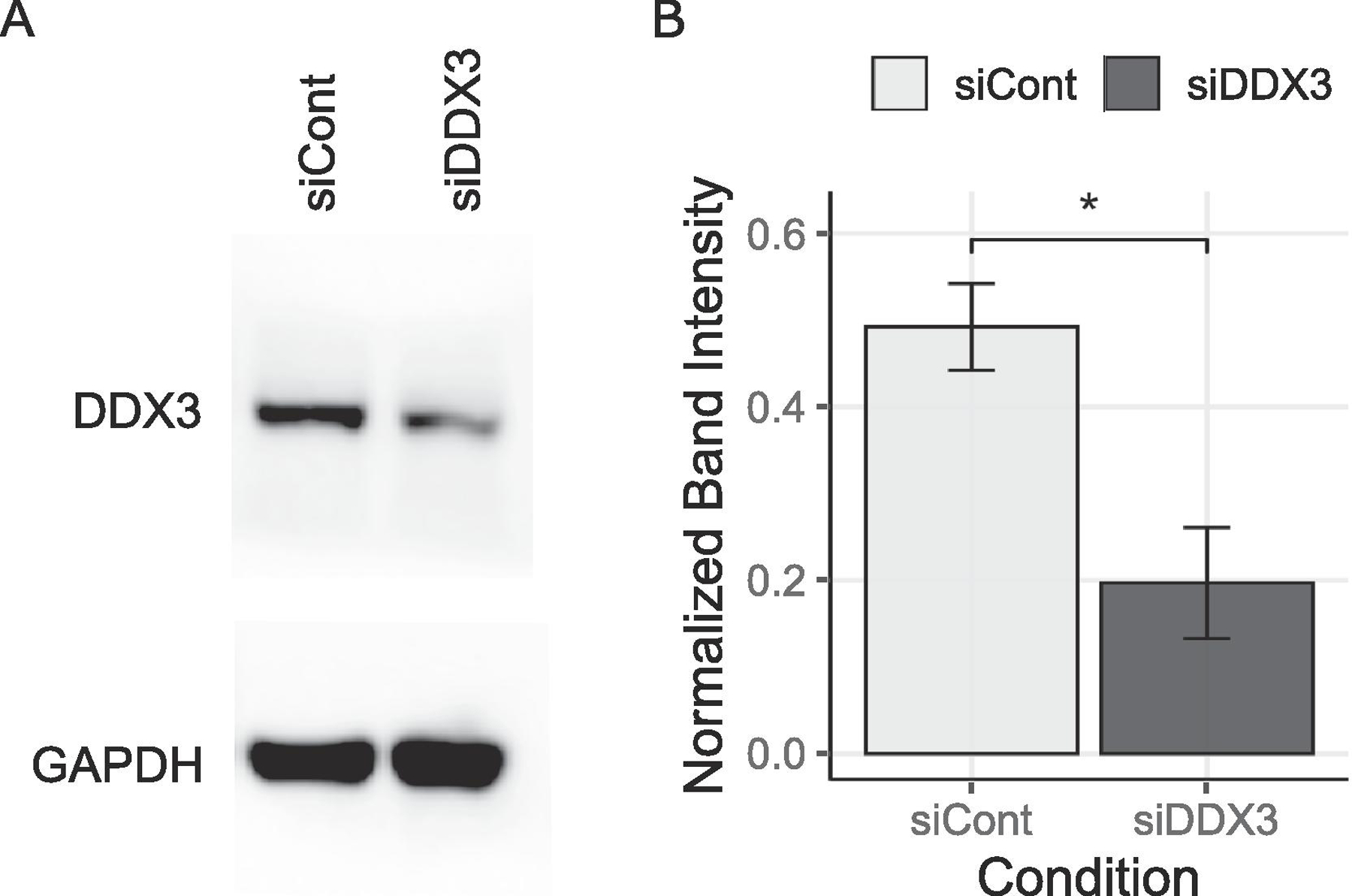
Quantification of DDX3 siRNA knockdown. (A) Western blot test using DDX3 antibody was quantified (B) by normalizing against GAPDH antibody. The T-Test was used to calculate statistical significance (* p < 0.05).
Without arsenite stress, the control and DDX3-depleted cultures show a similar G3BP1-positive aggregation density plot (Fig. 5C, 0 min). At 0 min, both the density plots and the median integrated intensities within the boxplots showed overlap between siCont and siDDX3 (Fig. 5C, 0 min). However, upon arsenite stress, the difference in G3BP1-positive aggregation behavior became distinct between the two siRNA treatment groups (Fig. 5C–D, 30–90 min). Compared with siCont SG assembly in light red, siDDX3 SG assembly in dark red showed a leftward shift for both density plots and boxplot profiles, indicating reductions in SG assembly (Fig. 5C–D). For all experimental time points, the peaks of siDDX3 in the density plot as well as the median integrated intensities for siDDX3 were lower than those of siCont (Fig. 5C–D, 30–90 min). This difference of SG integrated intensities between siCont and siDDX3 siRNA conditions indicates attenuation of SG assembly when DDX3 was depleted. Throughout three different time durations of arsenite treatment, DDX3-silenced cells formed significantly fewer G3BP1 positive SGs compared with the control scrambled siRNA, recapitulating the previously observed pharmacological effect of inhibiting DDX3.
Similar to the effects of RK-33, the SG quantification data showed DDX3 knockdown had no obvious effect on SG disassembly; the size and intensity of aggregated G3BP1 SG nucleations were comparable between siCont and siDDX3 (Fig. 5A–B). In this case, siDDX3 cultures showed greater median integrated intensities in 30 and 90 min conditions compared to those of siCont values, however, the differences were modest when compared to changes in SG assembly observed (Fig. 5F). In conclusion, the reduction of DDX3 mRNA attenuates DDX3-sequestered SG assembly but does not affect its disassembly.
3.5. Effects of the overexpression of DDX3 variants on SG assembly
We have determined that the SG assembly can be attenuated by depleting DDX3 with siRNA or pharmacologically inhibiting DDX3 ATPase and helicase activity. Next, we tested whether exogenously expressing wild-type or enzymatically inactive DDX3 variants affect SG assembly. U2OS cells were transiently transfected with the constructs expressing DDX3 wild-type and the DDX3 variants with abrogated ATPase and helicase site sequences. This was accomplished by targeting the DEAD motif with an E348Q mutation and targeting helicase activity with S382A and T384A substitutions [43]. All variants were fused with an N-terminal FLAG-tag. 48 h post-transfection cells were treated with arsenite for 90 min (Fig. 6). Interestingly, SG assembly was not affected by the overexpression of enzymatic-site mutant DDX3 variants (full arrows); rather, these cells formed SGs that were comparable to their un-transfected counterparts (arrowheads) (Fig. 6C–D). This observation was also true with DDX3 wild-type overexpression (Fig. 6B). The quantification of SGs and the analysis of these results did not reveal any significant differences in between wild-type and mutant DDX3 variants (data not shown). The vector control sample showed clear G3BP1-positive SGs, but not FLAG-positive granules, as expected (arrowheads, Fig. 6A). Taken together, we have observed that pharmacological inhibition or siRNA depletion decrease SG assembly, whereas overexpression of wild-type or DDX3 mutants did not produce observable effects in this experimental system.
Fig. 6.
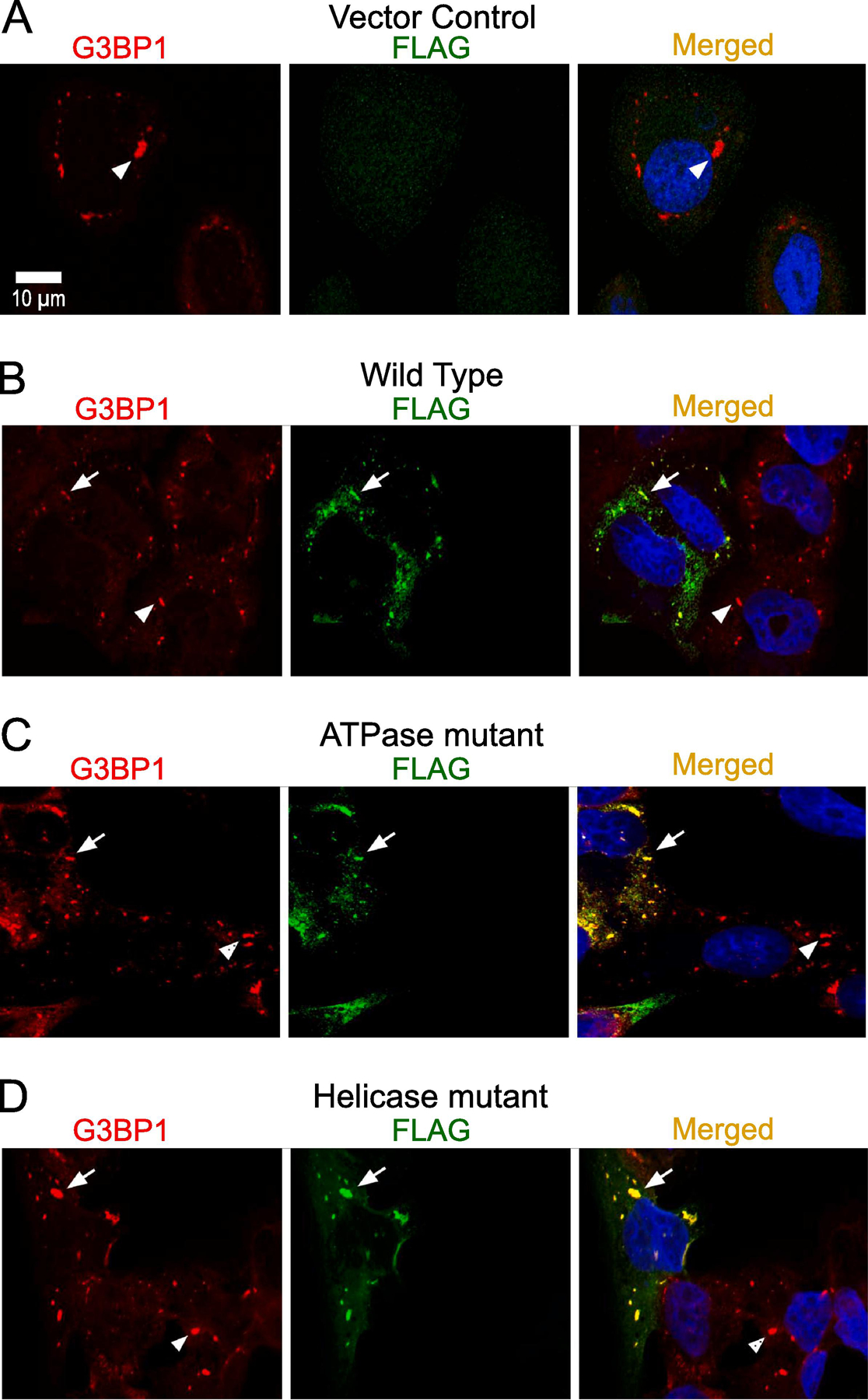
Transient expression of FLAG-DDX3 proteins. (A–D) Characterization of exogenous DDX3-FLAG on G3BP1-dependent SG assembly upon 90 min of 0.5 mM arsenite in U2OS cells with anti-G3BP1 (red) and anti-FLAG (green). Scale bar, 10 µm. (A) Vector control with G3BP1 and FLAG. Overexpressed (B) wild-type, (C) ATPase-site mutant, and (D) Helicase-site mutant with G3BP1 and FLAG. The full arrows represent localization of G3BP1 with overexpressed DDX3-FLAG; the arrowheads represent endogenous G3BP1-dependent SGs.
4. Discussion
Our study constitutes the analysis of the impact of DEAD-Box RNA Helicase 3 (DDX3) enzymatic activity on the dynamics of SG assembly and disassembly utilizing the DDX3 inhibitors RK-33 and 16D. Here, we find that pharmacological inhibition or siRNA knockdown of DDX3 suppresses SG assembly, but has only a marginal effect on SG disassembly.
We used a well-established SG induction system, and triggered oxidative stress in U2OS cells via arsenite. Time-course analyses of SG intensity in control and drug pre-treated cells subjected to arsenite determined that RK-33 and 16D inhibit SG assembly, which was mirrored by the effects of DDX3 knockdown. The magnitude of the reduction of SG formation by RK-33, which inhibits the ATPase activity of DDX3 was much more pronounced than the reduction in SG formation caused by D16, which only inhibits the helicase activity. The DDX3 protein dimerizes and binds dsRNA in its ATP-associated form, and the resulting conformational change triggers its helicase activity and dsRNA unwinding [24]. RK-33 might more effectively prevent DDX3-RNA interactions and therefore could confer a more pronounced effect on SG assembly than inhibiting DDX3 helicase activity with 16D. Additionally, inhibition of the ATPase activity may perturb ATP-dependent interactions of DDX3 with other proteins, which may not be affected by the helicase inhibitor [42].
Our results show that the enzymatic activities of DDX3 are much less involved in the SG disassembly. The time-course analyses of SG decondensation upon withdrawal from arsenite shows only minor differences between the control and the drug-treated or DDX3-depleted cells.
This finding better defines the role that DDX3’s enzymatic activity plays in SG initiation. DDX3, a ubiquitous RNA-binding protein, forms phase-separated droplets in vitro [44], and has been shown to promote phase separation when bound by ATP. Ribonucleoproteins (RNPs), protein-protein, and trans-RNA-RNA interactions all contribute to the assembly of the SGs [45, 46]. Since DDX3 preferentially functions in the unwinding of longer 5’ Untranslated Regions (UTRs) of mRNAs [47–50], mRNAs sequestered in SGs with long unwound 5’ UTRs could kinematically increase possible interfacing sites and surfaces for trans-RNA-RNA and RNA-protein interactions within the SGs, which may boost the kinetics of SG assembly (Fig. 7). A recent study has shown the involvement of mRNAs, freed from polysomes due to stress, contributing to phase-separation through binding to G3BP as well as the preferential recruitment of unstructured RNAs to the SGs through their enhanced associations with G3BP [19]. Although a functional DDX3 will unwind RNA secondary structures, a pharmacologically inhibited DDX3 will leave the long and complex 5’ UTRs intact. This could decrease the number of docking sites available for SG assembly kinematic interactions, leading to comparative attenuation of phase-separation and SG assembly. Contrary to the results with the ATPase inactive Ded1p helicase [51], the pharmacological inhibition of DDX3 ATPase activity does not increase the condensation of DDX3 positive structures, DDX3 localization in SGs, or SGs disassembly. Therefore, our results summarized in Fig. 7, suggest that the enzymatic activity of DDX3 regulates stress-induced assembly of SG. Our experimental data and the model (Fig. 7) corroborate with the recent finding that the stress-induced phase separation modulated by the DDX3 yeast homolog, Ded1p, results in the accumulation of mRNAs that contain complex 5’ UTRs in the SGs, while stress-induced transcripts with shorter 5’ UTRs escape the SGs and translational suppression [45, 52].
Fig. 7.
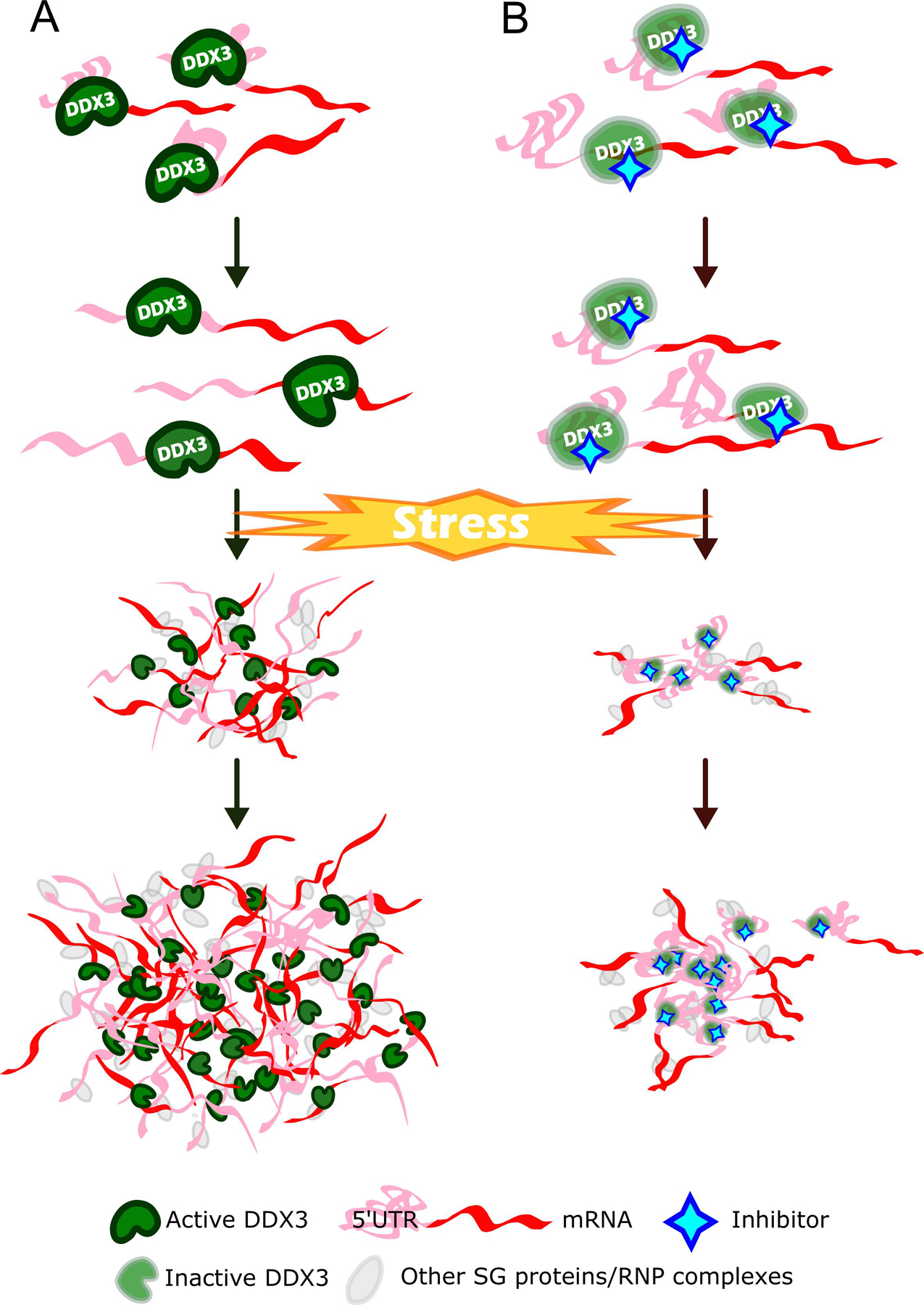
A schematic showing the effect of DDX3 inhibition on SG assembly. (A) DDX3 in its physiological state. Upon stress, mRNAs along with active DDX3s colocalize to SGs. These active DDX3s will unwind mRNAs with complex secondary structures of 5' UTR. This increases interactive surfaces for trans-RNA-RNA and RNA-protein intermolecular interactions, facilitating SG maturation. (B) DDX3 subjected to a pharmacological inhibition. Upon stress, mRNAs along with inactive DDX3s colocalize to SGs. However, the complex 5' UTRs of mRNAs will not be resolved by inactive DDX3s, resulting in less interactive surfaces for trans-RNA-RNA and RNA-protein intermolecular interactions, attenuating SG assembly.
Our results showing that the enzymatic activities of DDX3 contribute to SG assembly, rather than disassembly, fit the recent data suggesting a greater presence of promiscuous trans-RNA-RNA interactions in the SGs, which not only involves canonical Watson-Crick base pairing, but also involves a significant amount of non-canonical base pairing such as G quadraplex and Hoogstein base pairing [53, 54]. Moreover, interactions of RNA-binding proteins such as G3BP through homo and hetero oligomerization drive the condensation and stabilization of SGs [16, 19, 52]. These data support our findings that DDX3 enzymatic activity inhibition and siRNA knockdown have limited effects on the SG disassembly, since DDX3 may affect neither non-Watson-Crick interactions of RNAs nor RNP-based interactions.
Our study uncovers a new avenue for clinical implementation and development of DDX3-specific therapy targeting SG assembly. However, many questions remain prior to successful applications of such therapy. For example, DDX3 activity is involved in the development of Amyotrophic Lateral Sclerosis [55] and HIV-Associated Neurocognitive Disorders [31], while the inhibition of DDX3 is linked to Fragile X-Associated Tremor/Ataxia Syndrome [47], whereas mutations or deletions of DDX3X accounts for unexplained intellectual disabilities [21]. Moreover, DDX3 inhibitors have shown a strong promise in cancer treatments [25, 56, 57], yet, properly functional DDX3X is found to be a suppressor of medulloblastoma [58].
SGs hold a great impact on cell survival against a spectrum of ailments, from viral infection to tumor resilience and neurodegeneration. A concerted effort has been made to elucidate SG biochemical pathways and molecular processes. However, pharmacological intervention of SG dynamics has not yet been extensively assessed, and the effect of drug treatment on SG dynamics remains unclear. Identification of specific molecules that can mediate SG assembly could facilitate the development of novel therapeutics and may enhance responses to current therapies [13]. The effects of DDX3 enzymatic activities on the formation of pathological SGs need to be tested along with the applicability of the DDX3 inhibition for the treatment of SG-associated pathological states. Furthermore, proteomic approaches can be used to examine the effects of pharmacological interference on the composition of SGs.
To conclude, both DDX3 and SGs are implicated in many of the same diseases; therefore, modulating SG assembly through pharmacological inhibition of DDX3, and comparing inhibitors of DDX3 in vitro, could provide valuable screening tools for the evaluation of the developing DDX3 inhibitors [59]. Collectively, the results of this study reveal a crucial first glimpse into how pharmacological compounds could influence SG dynamics, and that SGs can be modulated by specific enzymatic inhibitors of DDX3.
Acknowledgements
We thank Drs. Giovanni Maga and Maurizio Botta from University of Siena, Italy, for their generous donation of 16D compound. We thank Drs Anna Kashina and Paula A. Vasquez for fruitful discussions and critical reading of the manuscript. We thank COBRE Center for Targeted Therapeutics Microscopy and Flow cytometry Core for image analysis. The work was supported by awards from NIH NIDA R21DA047936, R03DA043428 (MS), NIH CA223956 (MDW) and NIH NIGMS COBRE P20GM109091 (EB).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Competing Interest
None of the authors have any conflict of interest.
References
- [1].Corbet GA, Parker R, RNP Granule Formation: Lessons from P-Bodies and Stress Granules, Cold Spring Harb Symp Quant Biol (2020). [DOI] [PubMed]
- [2].Anderson P, Kedersha N, Ivanov P, Stress granules, P-bodies and cancer, Biochim Biophys Acta 1849(7) (2015) 861–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Markmiller S, Soltanieh S, Server KL, Mak R, Jin W, Fang MY, Luo EC, Krach F, Yang D, Sen A, Fulzele A, Wozniak JM, Gonzalez DJ, Kankel MW, Gao FB, Bennett EJ, Lecuyer E, Yeo GW, Context-Dependent and Disease-Specific Diversity in Protein Interactions within Stress Granules, Cell 172(3) (2018) 590–604 e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Lloyd RE, How do viruses interact with stress-associated RNA granules?, PLoS Pathog 8(6) (2012) e1002741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Anderson P, Kedersha N, Stressful initiations, J Cell Sci 115(Pt 16) (2002) 3227–34. [DOI] [PubMed] [Google Scholar]
- [6].Panas MD, Ivanov P, Anderson P, Mechanistic insights into mammalian stress granule dynamics, J Cell Biol 215(3) (2016) 313–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Ivanov P, Kedersha N, Anderson P, Stress Granules and Processing Bodies in Translational Control, Cold Spring Harb Perspect Biol 11(5) (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Wolozin B, Regulated protein aggregation: stress granules and neurodegeneration, Mol Neurodegener 7 (2012) 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Amorim R, Temzi A, Griffin BD, Mouland AJ, Zika virus inhibits eIF2alpha-dependent stress granule assembly, PLoS Negl Trop Dis 11(7) (2017) e0005775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Protter DS, Parker R, Principles and Properties of Stress Granules, Trends in cell biology 26(9) (2016) 668–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Gaete-Argel A, Marquez CL, Barriga GP, Soto-Rifo R, Valiente-Echeverria F, Strategies for Success. Viral Infections and Membraneless Organelles, Front Cell Infect Microbiol 9 (2019) 336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ryan VH, Fawzi NL, Physiological, Pathological, and Targetable Membraneless Organelles in Neurons, Trends Neurosci 42(10) (2019) 693–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Sahoo PK, Lee SJ, Jaiswal PB, Alber S, Kar AN, Miller-Randolph S, Taylor EE, Smith T, Singh B, Ho TS, Urisman A, Chand S, Pena EA, Burlingame AL, Woolf CJ, Fainzilber M, English AW, Twiss JL, Axonal G3BP1 stress granule protein limits axonal mRNA translation and nerve regeneration, Nat Commun 9(1) (2018) 3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Tauber D, Tauber G, Parker R, Mechanisms and Regulation of RNA Condensation in RNP Granule Formation, Trends Biochem Sci (2020). [DOI] [PMC free article] [PubMed]
- [15].Nunes C, Mestre I, Marcelo A, Koppenol R, Matos CA, Nobrega C, MSGP: the first database of the protein components of the mammalian stress granules, Database : the journal of biological databases and curation 2019 (2019). [DOI] [PMC free article] [PubMed]
- [16].Sanders DW, Kedersha N, Lee DSW, Strom AR, Drake V, Riback JA, Bracha D, Eeftens JM, Iwanicki A, Wang A, Wei MT, Whitney G, Lyons SM, Anderson P, Jacobs WM, Ivanov P, Brangwynne CP, Competing Protein-RNA Interaction Networks Control Multiphase Intracellular Organization, Cell 181(2) (2020) 306–324 e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kedersha N, Panas MD, Achorn CA, Lyons S, Tisdale S, Hickman T, Thomas M, Lieberman J, McInerney GM, Ivanov P, Anderson P, G3BP-Caprin1-USP10 complexes mediate stress granule condensation and associate with 40S subunits, J Cell Biol 212(7) (2016) 845–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Ash PE, Vanderweyde TE, Youmans KL, Apicco DJ, Wolozin B, Pathological stress granules in Alzheimer’s disease, Brain Res 1584 (2014) 52–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Guillen-Boixet J, Kopach A, Holehouse AS, Wittmann S, Jahnel M, Schlussler R, Kim K, Trussina I, Wang J, Mateju D, Poser I, Maharana S, Ruer-Gruss M, Richter D, Zhang X, Chang YT, Guck J, Honigmann A, Mahamid J, Hyman AA, Pappu RV, Alberti S, Franzmann TM, RNA-Induced Conformational Switching and Clustering of G3BP Drive Stress Granule Assembly by Condensation, Cell 181(2) (2020) 346–361 e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Samir P, Kesavardhana S, Patmore DM, Gingras S, Malireddi RKS, Karki R, Guy CS, Briard B, Place DE, Bhattacharya A, Sharma BR, Nourse A, King SV, Pitre A, Burton AR, Pelletier S, Gilbertson RJ, Kanneganti T-D, DDX3X acts as a live-or-die checkpoint in stressed cells by regulating NLRP3 inflammasome, Nature (2019). [DOI] [PMC free article] [PubMed]
- [21].Lennox AL, Hoye ML, Jiang R, Johnson-Kerner BL, Suit LA, Venkataramanan S, Sheehan CJ, Alsina FC, Fregeau B, Aldinger KA, Moey C, Lobach I, Afenjar A, Babovic-Vuksanovic D, Bezieau S, Blackburn PR, Bunt J, Burglen L, Campeau PM, Charles P, Chung BHY, Cogne B, Curry C, D’Agostino MD, Di Donato N, Faivre L, Heron D, Innes AM, Isidor B, Keren B, Kimball A, Klee EW, Kuentz P, Kury S, Martin-Coignard D, Mirzaa G, Mignot C, Miyake N, Matsumoto N, Fujita A, Nava C, Nizon M, Rodriguez D, Blok LS, Thauvin-Robinet C, Thevenon J, Vincent M, Ziegler A, Dobyns W, Richards LJ, Barkovich AJ, Floor SN, Silver DL, Sherr EH, Pathogenic DDX3X Mutations Impair RNA Metabolism and Neurogenesis during Fetal Cortical Development, Neuron 106(3) (2020) 404–420 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Fazi R, Tintori C, Brai A, Botta L, Selvaraj M, Garbelli A, Maga G, Botta M, Homology Model-Based Virtual Screening for the Identification of Human Helicase DDX3 Inhibitors, J Chem Inf Model 55(11) (2015) 2443–54. [DOI] [PubMed] [Google Scholar]
- [23].Raj S, Bagchi D, Orero JV, Banroques J, Tanner NK, Croquette V, Mechanistic characterization of the DEAD-box RNA helicase Ded1 from yeast as revealed by a novel technique using single-molecule magnetic tweezers, Nucleic Acids Res 47(7) (2019) 3699–3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Song H, Ji X, The mechanism of RNA duplex recognition and unwinding by DEAD-box helicase DDX3X, Nat Commun 10(1) (2019) 3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Bol GM, Vesuna F, Xie M, Zeng J, Aziz K, Gandhi N, Levine A, Irving A, Korz D, Tantravedi S, Heerma van Voss MR, Gabrielson K, Bordt EA, Polster BM, Cope L, van der Groep P, Kondaskar A, Rudek MA, Hosmane RS, van der Wall E, van Diest PJ, Tran PT, Raman V, Targeting DDX3 with a small molecule inhibitor for lung cancer therapy, EMBO Mol Med 7(5) (2015) 648–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Lai MC, Sun HS, Wang SW, Tarn WY, DDX3 functions in antiviral innate immunity through translational control of PACT, FEBS J 283(1) (2016) 88–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Adjibade P, Grenier St-Sauveur V, Bergeman J, Huot ME, Khandjian EW, Mazroui R, DDX3 regulates endoplasmic reticulum stress-induced ATF4 expression, Sci Rep 7(1) (2017) 13832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Chen HH, Yu HI, Tarn WY, DDX3 Modulates Neurite Development via Translationally Activating an RNA Regulon Involved in Rac1 Activation, J Neurosci 36(38) (2016) 9792–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Chen HH, Yu HI, Yang MH, Tarn WY, DDX3 Activates CBC-eIF3-Mediated Translation of uORF-Containing Oncogenic mRNAs to Promote Metastasis in HNSCC, Cancer Res 78(16) (2018) 4512–4523. [DOI] [PubMed] [Google Scholar]
- [30].Chao CH, Chen CM, Cheng PL, Shih JW, Tsou AP, Lee YH, DDX3, a DEAD box RNA helicase with tumor growth-suppressive property and transcriptional regulation activity of the p21waf1/cip1 promoter, is a candidate tumor suppressor, Cancer Res 66(13) (2006) 6579–88. [DOI] [PubMed] [Google Scholar]
- [31].Aksenova M, Sybrandt J, Cui B, Sikirzhytski V, Ji H, Odhiambo D, Lucius MD, Turner JR, Broude E, Pena E, Lizarraga S, Zhu J, Safro I, Wyatt MD, Shtutman M, Inhibition of the Dead Box RNA Helicase 3 Prevents HIV-1 Tat and Cocaine-Induced Neurotoxicity by Targeting Microglia Activation, J Neuroimmune Pharmacol (2019) 591438. [DOI] [PMC free article] [PubMed]
- [32].Frohlich A, Rojas-Araya B, Pereira-Montecinos C, Dellarossa A, Toro-Ascuy D, Prades-Perez Y, Garcia-de-Gracia F, Garces-Alday A, Rubilar PS, Valiente-Echeverria F, Ohlmann T, Soto-Rifo R, DEAD-box RNA helicase DDX3 connects CRM1-dependent nuclear export and translation of the HIV-1 unspliced mRNA through its N-terminal domain, Biochim Biophys Acta 1859(5) (2016) 719–30. [DOI] [PubMed] [Google Scholar]
- [33].Brai A, Fazi R, Tintori C, Zamperini C, Bugli F, Sanguinetti M, Stigliano E, Este J, Badia R, Franco S, Martinez MA, Martinez JP, Meyerhans A, Saladini F, Zazzi M, Garbelli A, Maga G, Botta M, Human DDX3 protein is a valuable target to develop broad spectrum antiviral agents, Proc Natl Acad Sci U S A 113(19) (2016) 5388–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Su CY, Lin TC, Lin YF, Chen MH, Lee CH, Wang HY, Lee YC, Liu YP, Chen CL, Hsiao M, DDX3 as a strongest prognosis marker and its downregulation promotes metastasis in colorectal cancer, Oncotarget 6(21) (2015) 18602–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Oliver D, Ji H, Liu P, Gasparian A, Gardiner E, Lee S, Zenteno A, Perinskaya LO, Chen M, Buckhaults P, Broude E, Wyatt MD, Valafar H, Pena E, Shtutman M, Identification of novel cancer therapeutic targets using a designed and pooled shRNA library screen, Sci Rep 7 (2017) 43023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Shih JW, Wang WT, Tsai TY, Kuo CY, Li HK, Wu Lee YH, Critical roles of RNA helicase DDX3 and its interactions with eIF4E/PABP1 in stress granule assembly and stress response, Biochem J 441(1) (2012) 119–29. [DOI] [PubMed] [Google Scholar]
- [37].Matheny T, Rao BS, Parker R, Transcriptome-Wide Comparison of Stress Granules and P-Bodies Reveals that Translation Plays a Major Role in RNA Partitioning, Mol Cell Biol 39(24) (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fritzler MJ, Scheuner D, Kaufman RJ, Golan DE, Anderson P, Stress granules and processing bodies are dynamically linked sites of mRNP remodeling, J Cell Biol 169(6) (2005) 871–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Takahashi M, Higuchi M, Matsuki H, Yoshita M, Ohsawa T, Oie M, Fujii M, Stress granules inhibit apoptosis by reducing reactive oxygen species production, Mol Cell Biol 33(4) (2013) 815–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Fujimura K, Katahira J, Kano F, Yoneda Y, Murata M, Microscopic dissection of the process of stress granule assembly, Biochimica et biophysica acta 1793(11) (2009) 1728–37. [DOI] [PubMed] [Google Scholar]
- [41].Wheeler JR, Matheny T, Jain S, Abrisch R, Parker R, Distinct stages in stress granule assembly and disassembly, Elife 5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Çelik H, Sajwan KP, Selvanathan SP, Marsh BJ, Pai AV, Kont YS, Han J, Minas TZ, Rahim S, Erkizan HV, Toretsky JA, Üren A, Ezrin Binds to DEAD-Box RNA Helicase DDX3 and Regulates Its Function and Protein Level, Molecular and Cellular Biology 35(18) (2015) 3145–3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Tanner NK, Linder P, DExD/H box RNA helicases: from generic motors to specific dissociation functions, Mol Cell 8(2) (2001) 251–62. [DOI] [PubMed] [Google Scholar]
- [44].Hondele M, Sachdev R, Heinrich S, Wang J, Vallotton P, Fontoura BMA, Weis K, DEAD-box ATPases are global regulators of phase-separated organelles, Nature 573(7772) (2019) 144–148. [DOI] [PubMed] [Google Scholar]
- [45].Iserman C, Desroches Altamirano C, Jegers C, Friedrich U, Zarin T, Fritsch AW, Mittasch M, Domingues A, Hersemann L, Jahnel M, Richter D, Guenther UP, Hentze MW, Moses AM, Hyman AA, Kramer G, Kreysing M, Franzmann TM, Alberti S, Condensation of Ded1p Promotes a Translational Switch from Housekeeping to Stress Protein Production, Cell 181(4) (2020) 818–831 e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Khong A, Matheny T, Jain S, Mitchell SF, Wheeler JR, Parker R, The Stress Granule Transcriptome Reveals Principles of mRNA Accumulation in Stress Granules, Mol Cell 68(4) (2017) 808–820 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Linsalata AE, He F, Malik AM, Glineburg MR, Green KM, Natla S, Flores BN, Krans A, Archbold HC, Fedak SJ, Barmada SJ, Todd PK, DDX3X and specific initiation factors modulate FMR1 repeat-associated non-AUG-initiated translation, EMBO Rep (2019) e47498. [DOI] [PMC free article] [PubMed]
- [48].Ku YC, Lai MH, Lo CC, Cheng YC, Qiu JT, Tarn WY, Lai MC, DDX3 Participates in Translational Control of Inflammation Induced by Infections and Injuries, Mol Cell Biol (2018). [DOI] [PMC free article] [PubMed]
- [49].Jankowsky E, Guenther UP, A helicase links upstream ORFs and RNA structure, Current genetics (2018). [DOI] [PMC free article] [PubMed]
- [50].Guenther UP, Weinberg DE, Zubradt MM, Tedeschi FA, Stawicki BN, Zagore LL, Brar GA, Licatalosi DD, Bartel DP, Weissman JS, Jankowsky E, The helicase Ded1p controls use of near-cognate translation initiation codons in 5’ UTRs, Nature 559(7712) (2018) 130–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Hondele M, Sachdev R, Heinrich S, Wang J, Vallotton P, Fontoura BMA, Weis K, DEAD-box ATPases are global regulators of phase-separated organelles, Nature (2019). [DOI] [PubMed]
- [52].Singh J, Phase Separation of RNA Helicase Triggers Stress-Responsive Translational Switch, Trends Biochem Sci (2020). [DOI] [PubMed]
- [53].Van Treeck B, Protter DSW, Matheny T, Khong A, Link CD, Parker R, RNA self-assembly contributes to stress granule formation and defining the stress granule transcriptome, Proc Natl Acad Sci U S A 115(11) (2018) 2734–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Leontis NB, Stombaugh J, Westhof E, The non-Watson-Crick base pairs and their associated isostericity matrices, Nucleic Acids Res 30(16) (2002) 3497–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Chen Y, Wang Q, Wang Q, Liu H, Zhou F, Zhang Y, Yuan M, Zhao C, Guan Y, Wang X, DDX3 binding with CK1ε was closely related to motor neuron degeneration of ALS by affecting neurite outgrowth, American Journal of Translational Research 9(10) (2017) 4627–4639. [PMC free article] [PubMed] [Google Scholar]
- [56].Riva V, Maga G, From the magic bullet to the magic target: exploiting the diverse roles of DDX3X in viral infections and tumorigenesis, Future medicinal chemistry (2019). [DOI] [PubMed]
- [57].Bol GM, Xie M, Raman V, DDX3, a potential target for cancer treatment, Molecular Cancer 14(1) (2015) 188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Patmore DM, Jassim A, Nathan E, Gilbertson RJ, Tahan D, Hoffmann N, Tong Y, Smith KS, Kanneganti TD, Suzuki H, Taylor MD, Northcott P, Gilbertson RJ, DDX3X Suppresses the Susceptibility of Hindbrain Lineages to Medulloblastoma, Dev Cell (2020). [DOI] [PMC free article] [PubMed]
- [59].Nakao S, Nogami M, Iwatani M, Imaeda T, Ito M, Tanaka T, Tawada M, Endo S, Cary DR, Ohori M, Imaeda Y, Kawamoto T, Aparicio S, Nakanishi A, Araki S, Identification of a selective DDX3X inhibitor with newly developed quantitative high-throughput RNA helicase assays, Biochem Biophys Res Commun 523(3) (2020) 795–801. [DOI] [PubMed] [Google Scholar]


