Fig. 5.
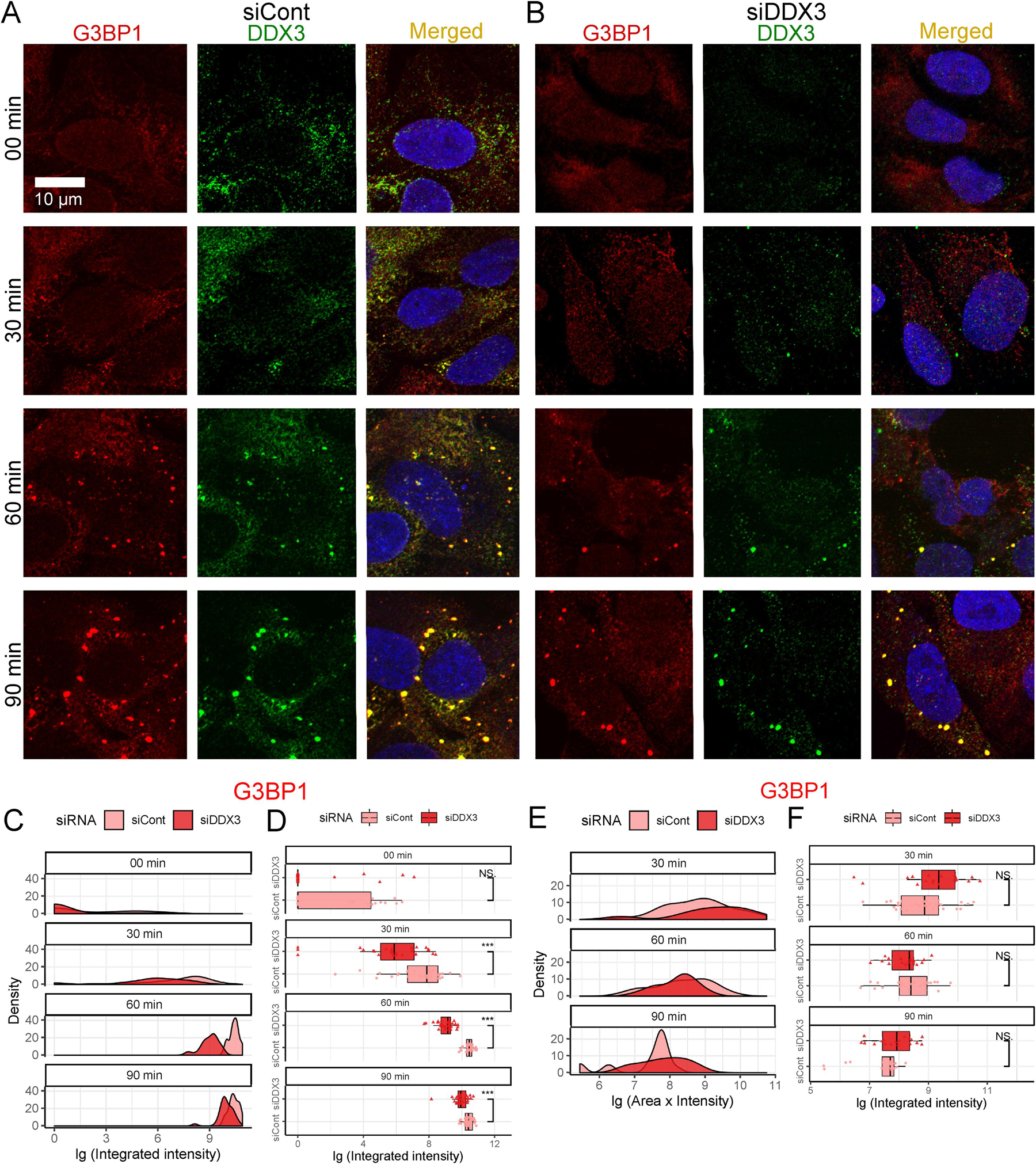
Effects of DDX3 siRNA knockdown on SG assembly and disassembly. (A–B) SG assembly was examined by Immunofluorescence with anti-G3BP1 (red) and anti-DDX3 (green). Scale bar, 10 µm. Cells subjected to 00 min, 30 min, 60 min, and 90 min of 0.5 mM arsenite stress either (A) without or (B) with DDX3 siRNA knockdown are quantified in (C and D). Cells were subjected to 60 min of 0.5 mM arsenite treatment followed by replaceing with fresh media to recover for 30 min, 60 min, and 90 min without or with DDX3 siRNA knockdown and are quantified in (E and F). (C–F) Time course distributions of G3BP1 granules either without (lighter red) or with (darker red) DDX3 siRNA knockdown. (C and E) shows density plots and (D and F) shows box-and-whisker plots. The boxes cover 50% of data, and lines within boxes indicate median values. The Wilcoxon Signed-Rank Test was used to calculate statistical significance (* p < 0.05; ** p < 0.01; *** p < 0.001).
