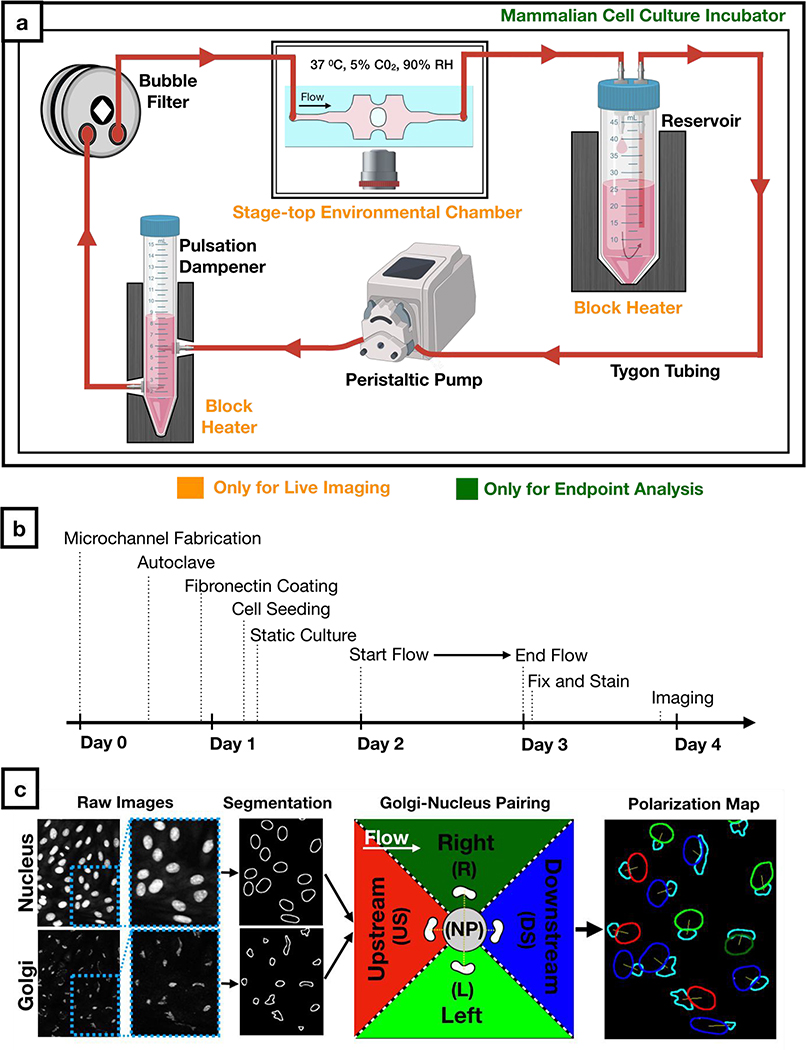
Figure 3: Workflow for cell culture experiments and image analysis.
a) Schematic of the sterile flow circuit used with the microfluidic chip. The flow circuit can be set up on a microscope stage for live imaging under flow or it can be placed in a cell culture incubator for endpoint analysis.
b) Experimental timeline for Golgi-nucleus morphology studies. Human umbilical vein endothelial cells (HUVECs) were seeded inside the microfluidic chip 16 hours before initiating flow and fixed after 24 hours of flow.
c) Fluorescent images of nuclei and Golgi were analyzed automatically using a custom-written macro to define and pair each nucleus and Golgi as well as to categorize each cell as nonpolarized (gray) or polarized. Polarized cells were binned into one of four Golgi-nucleus orientation categories: upstream (red), downstream (blue), right (dark green), left (light green), with respect to the direction of flow. Orientation maps outline Golgi (cyan) and nucleus, with nucleus color indicating the orientation of the cell.