Figure 4: Smoothened is ubiquitinated by the MEGF8-MGRN1 complex.
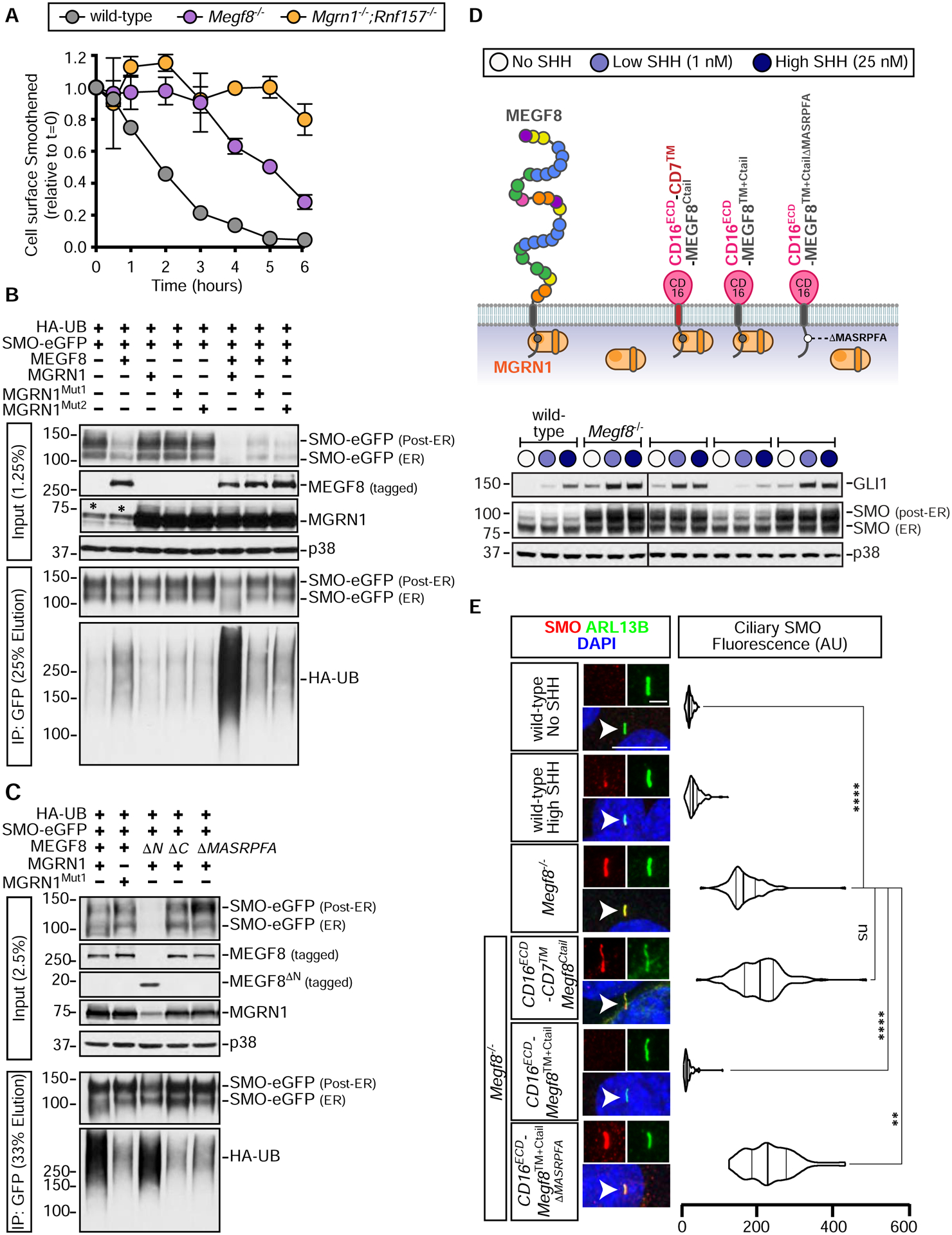
(A) Degradation of cell-surface SMO in NIH/3T3 cells of the indicated genotypes. See Fig. S5 for details. Error bars represent the standard error of two independent replicates.
(B and C) SMO ubiquitination was assessed after transient co-expression of the indicated proteins in HEK293T cells (see Fig. 3A). Cells were lysed under denaturing conditions, SMO was purified by IP, and the amount of HA-UB covalently conjugated to SMO assessed using immunoblotting with an anti-HA antibody. An asterisk (*) indicates endogenous MGRN1.
(D and E) Total GLI1 and SMO abundances were measured by immunoblotting (D) and ciliary SMO (n~50 cilia) by fluorescence confocal microscopy (E) in Megf8−/− cells expressing various CD16/CD7/MEGF8 chimeras. The ability of these chimeras to support SMO ubiquitination is shown in Fig. S6D and the abundances of chimeras at the cell surface is shown in Fig. S6E. Statistical significance in (E) was determined by the Kruskal-Wallis test; not-significant (ns) > 0.05, **p-value ≤ 0.01, and ****p-value ≤ 0.0001. Scale bars, 10 μm in merged panels and 2 μm in zoomed displays. See also Figures S5 and S6.
