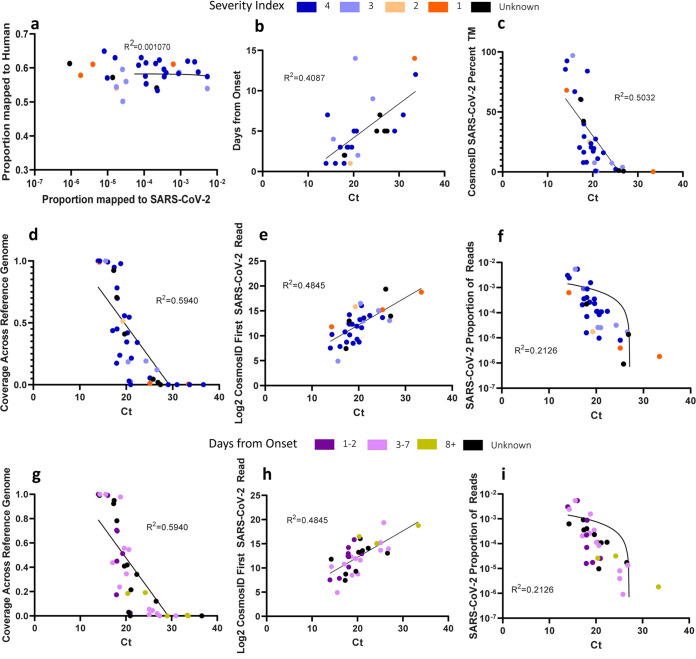
FIG 1.
Relationships of metatranscriptomic sequencing sample characteristics to sequencing results. (a) Relationship between the proportion of reads mapped to SARS-CoV-2 and the number of reads matched to human sequences. (b to i) Plots of sequencing results against the Ct values of metatranscriptomic sequencing samples. Panels a to f are color coded by severity index values, and panels g to i are color coded by days from onset of symptoms. The severity index was defined on a scale of 1 to 4, as follows: 4, not admitted; 3, admitted; 2, intensive care unit; and 1, required ventilator. Black dots represent samples with unknown onset or severity index values. (b) Relationship of Ct values determined by LDT-RT-PCR to the number of days from symptom onset. (c) Relationship between Ct values and the proportion of total matches of SARS-CoV-2 by CosmosID. (d, g) Relationship between Ct values and the sequencing coverage across the SARS-CoV-2 strain Hu-1 genome. (e, h) Relationship between Ct values and the number of reads analyzed by CosmosID until the first SARS-CoV-2 read was detected. (f, i) Relationship of Ct values to the proportion of SARS-CoV-2 reads present in the sample. Simple linear regression analysis was performed for each set, and the null hypothesis is rejected for panels b to i (P < 0.01), and for each set, R2 is reported.