Figure 2.
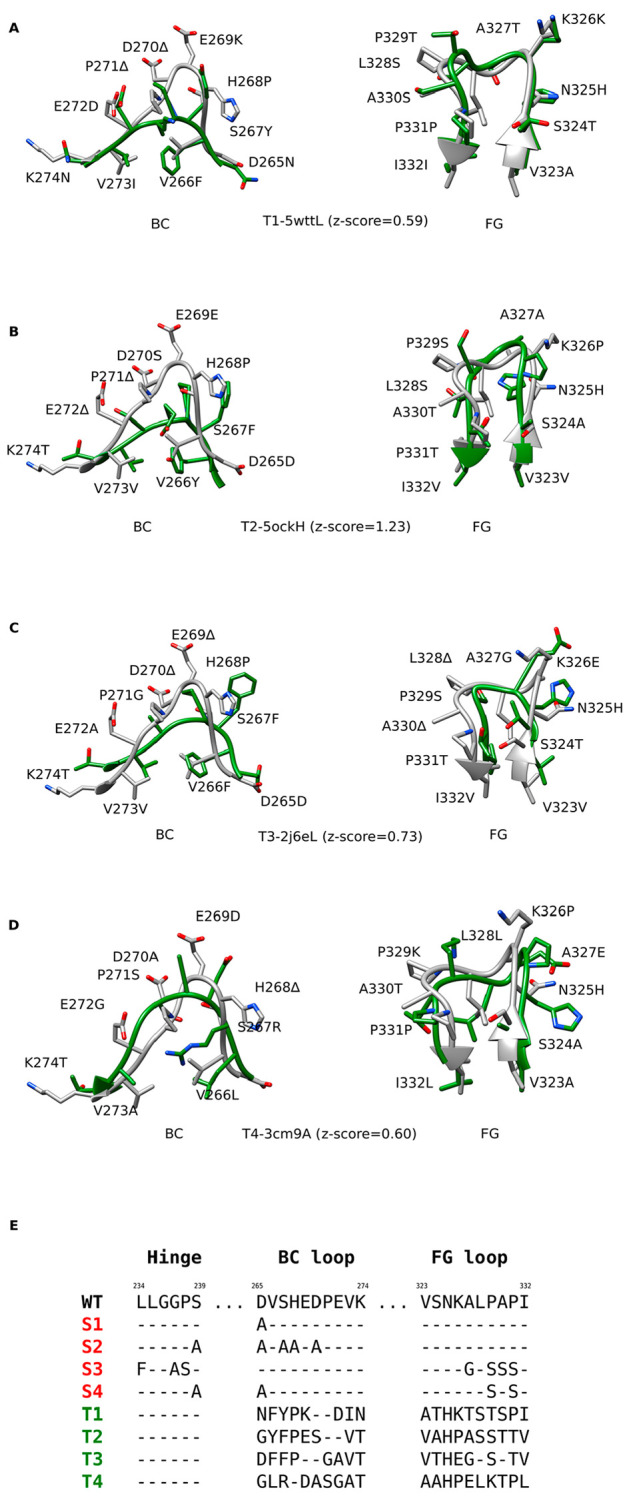
Designed Fc variants. A–D) Structural superpositions of the wild-type Fc region BC and FG loops (white) with the loops from different antibodies (green), based on which the loop grafted variants T1–T4 were constructed. Z-scores denote the degrees of local structural similarity between the binding site for FcγRIIIa on the Fc region and other antibody structures (PDB and Chain IDs) determined by ProBiS. E) Alignments of the wild-type (WT) amino acid sequence with the sequences of standard Fc variants (S1–S4, red) and the sequences of the loop grafted Fc variants (T1–T4, green). The sequence alignments for the latter were determined based on the structural alignments between the wild-type and the grafted loops.
