Figure 4. TFEB Target Genes Are Differentially Sensitive to Nuclear TFEB.
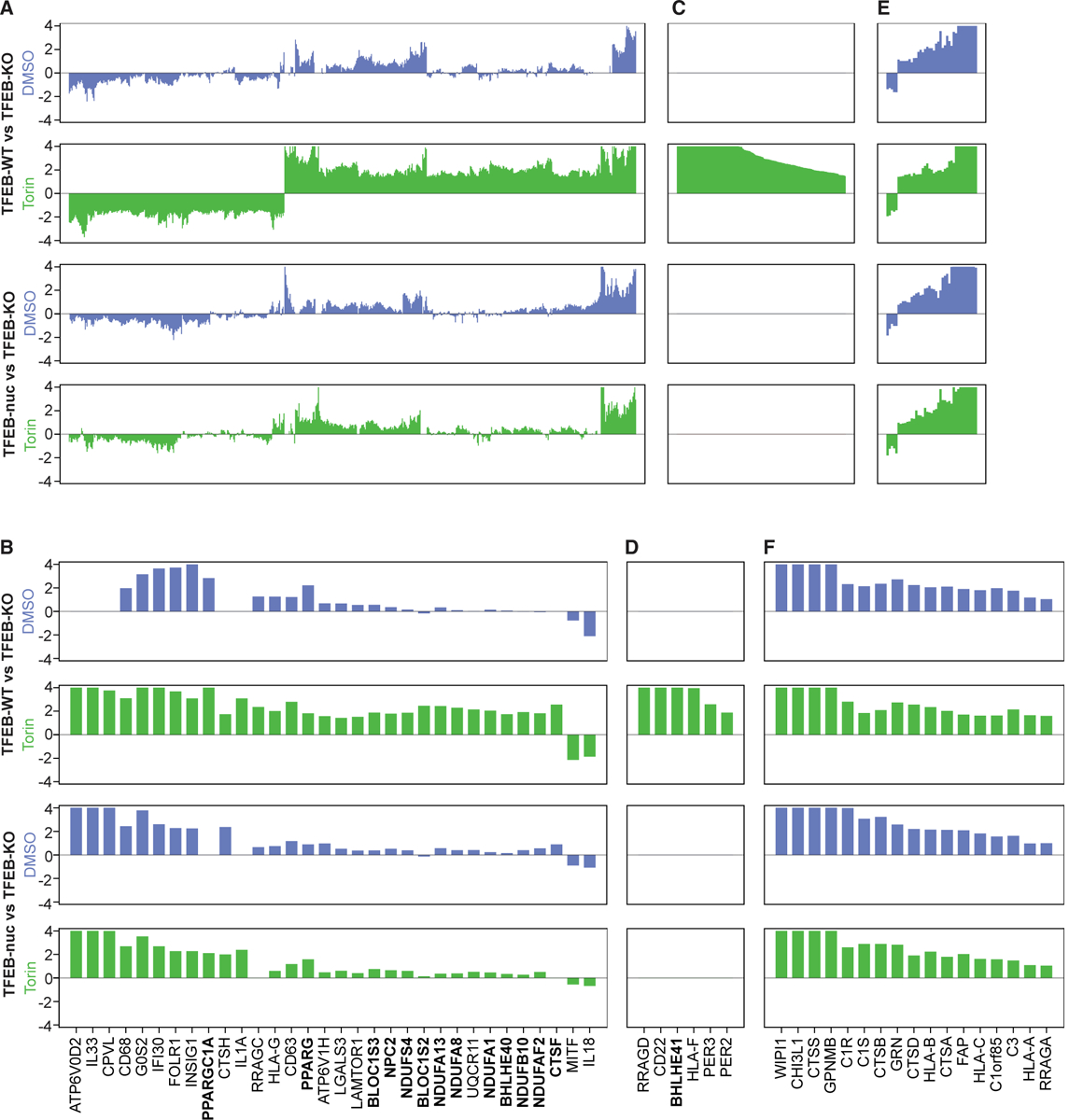
TFEB-KO, TFEB-WT, and TFEB-nuc cells were treated with Torin or DMSO and then processed for RNA sequencing analysis. Panels show differential gene expression from steady state (DMSO, blue) and Torin-treated (green) TFEB-WT versus TFEB-KO cells and TFEB-nuc versus TFEB-KO cells. Each bar corresponds to a gene, and the y axis represents logFC of differential gene expression (truncated logFC ± ln4). Genes represented in the bar plots are all genes (A, C, and E) or select genes (B, D, and F) with significant differential expression (logFC > ln4 or logFC < −ln4 and q < 0.01) in TFEB-WT relative to TFEB-KO cells following Torin treatment. For each differential expression comparison, (A) and (B) represent genes significantly upregulated with Torin treatment, (C) and (D) represents genes significantly upregulated with Torin treatment yet not transcribed at detectable levels without Torin stimulation, and (E) and (F) represent TFEB-dependent genes for which transcription did not significantly change in response to Torin stimulation. Genes in bold indicate those highlighted in Results. See also Figure S4 and Table S4.
