Abstract
The solute carrier family 52 member 3 (SLC52A3) gene encodes riboflavin transporter protein which is essential to maintain mitochondrial function in cells. In our research, we found that SLC52A3 rs13042395 C > T variation was significantly associated with poor survival in a 926 Chinese gastric cancer (GCa) patients cohort (CC/CT genotype versus TT genotype, HR = 0.57, 95%CI (0.40‐0.82), log‐rank P = 0.015). The SLC52A3 rs13042395 C > T change led to its increased mRNA expression according to expression quantitative trait loci analysis (P = 0.0029). In vitro, it was revealed that rs13042395 C allele had higher binding affinity to inhibitory transcription factor Meis homeobox 1 (MEIS1) compared with T allele, knock‐down of MEIS1 could up‐regulate SLC52A3, and overexpression of SLC52A3 contributed to the increased ability of proliferation, colony formation, migration and invasion in GCa cells. Subsequently, the bioinformatics analysis combined with experiments in vitro suggested that Gap junction protein alpha 1 (GJA1) was the downstream effector of SLC52A3, SLC52A3 may promote the GCa cells aggressiveness by down‐regulating the GJA1 expression. Overall, SLC52A3 genetic variant rs13042395 C > T change was associated with poorer survival in Chinese GCa patients and increased SLC52A3 expression by interaction with MEIS1. SLC52A3 promoted the GCa cells aggressiveness by down‐regulating the GJA1 expression.
Keywords: gastric cancer, prognosis, single nucleotide polymorphism, SLC52A3
1. INTRODUCTION
Over 679 000 patients suffered from gastric carcinoma (GCa) with an estimated nearly 500 000 GCa‐related deaths in 2015, making it the second most deadly cancer in China. 1 Despite the advance achieved in surgery, chemotherapy, radiotherapy and targeted therapies in the past few decades, the median overall survival (OS) of advanced GCa remained 10‐12 months. 2 Hence, it was critical to select poor survival GCa patients for earlier intervention which was promising to prolong the survival time of them.
Recently, several biomarkers were identified for the prognosis of GCa which was potentially able to select high‐risk patients, such as circulating non‐coding RNA and some somatic genes mutations. 3 , 4 Several germline mutations have already been investigated for the prognosis in GCa. 5 , 6 However, few of them further explored the molecular mechanism underlying the prognosis effect.
The solute carrier family 52 member 3 (SLC52A3) gene encodes the riboflavin transporter protein, which was crucial for the riboflavin absorption. 7 Previous studies suggested that SLC52A3 gene was highly abnormally expressed in oesophageal tumour tissue, and the depletion of SLC52A3 gene would significantly inhibit the proliferation of oesophageal carcinoma cells, 8 which indicated that SLC52A3 gene played an important role in the progression of cancer cells. Germline mutation (rs13042395 C > T) on SLC52A3 gene was identified as a risk locus for gastric cancer incidence by genome‐wide association studies, and it was found that rs13042395 C > T change significantly increased the risk of cancer incidence in various cancer types, notably in the GCa patients. 9 , 10 However, the molecular mechanism remained ambiguous and the research on the association of germline mutation in the SLC52A3 gene and the survival of GCa patients was hardly seen. In the present study, we investigated associations between genetic mutations of the SLC52A3 rs13042395 C > T change and survival of Chinese GCa patients and subsequently explored mechanistic basis of the observed associations.
2. METHOD AND MATERIAL
2.1. Study population
From January 2009 to March 2011, a total of 926 unrelated Han ethnic Chinese patients were recruited from Fudan University Shanghai Cancer Center (FUSCC) in Eastern China who was newly diagnosed or pathologically confirmed primary GCa. Peripheral blood samples of this GCa patients were provided by the tissue bank of FUSCC. All of the 926 patients had signed a written informed consent to donate their biological samples to the tissue bank for scientific research. Clinical information of each patients was collected. Our research proposal was approved by the FUSCC institutional review board.
2.2. Genotyping, quality control and survival analysis
DNA of study patients was extracted from peripheral blood cells. Single nucleotide polymorphisms (SNPs) were genotyped via a matrix‐assisted laser desorption/ionization time‐of‐flight (MALDI‐TOF) mass spectrometer using the MassARRAY Analyzer 4 platform (Sequenom, CA, USA). All the primers were designed by Assay Design Suite v2.0 from Mysequenom online software (www.mysequenom.com). The standard polymerase chain reaction (PCR) reaction was conducted in a total volume of 5 μL system containing 10 ng of genomic DNA. One negative control and one duplicate sample were used for quality controls in every 96‐well plates. Genotyping results of 5% patients were repeated, and the consistency was 100%.
A total of 926 patients were divided into two groups according to their rs13042395 genotype. Kaplan‐Meier (KM) curve was used to illustrate the difference and log‐rank test was performed to provide the statistics. Multivariate analysis was calculated by Cox proportional hazards regression which was adjusted by clinical variables. Stratification analysis was conducted to explore the interaction of the genetic variant with clinical variables.
2.3. Expression quantitative trait loci analysis and validation in vitro
Expression quantitative trait loci analysis was conducted using rs13042395 genotype and SLC52A3 mRNA expression data in normal gastric tissue from the Genotype‐Tissue Expression (GTEx) database. The different expression violin plot was directly available from the GTEx website (https://www.gtexportal.org/).
The Jaspar web server (http://jaspar.genereg.net/) was used to detect the putative transcription factors binding to the SLC52A3 rs13042395 allele. Subsequently, the binding affinity change resulted from rs13042395 C > T change was suggested. The Meis homeobox 1 (MEIS1) was found with the most significant binding affinity change, and MEIS1 then was selected for the further research.
The association of MEIS1 and SLC52A3 expression was assessed by GEPIA webserver (http://gepia.cancer‐pku.cn), and plot figure was obtained. Validation experiments were performed. Three types of siRNA targeting the different site of MEIS1 were purchased from the GenePharma (Shanghai China). These siRNA was used to transfect the BGC823 cells, respectively, using Lipofectamine™ RNAiMAX (Thermo Fisher, MA, Waltham, USA). siMEIS1‐1 sequence: CUGUCAAUGACGCUUUAAATT (forward); siMEIS1‐2 sequence: GCUCGUCAGAGUCAUUCAATT (forward); siMEIS1‐3 sequence: GCCUAUCGAUUUGGUGAUATT (forward). Western blotting and real‐time PCR were performed to detect the MEIS1 and SLC52A3 expression change.
2.4. Target sequencing and electrophoresis mobility shift assay
We sequenced the rs13042395 locus in BGC823, MGC803 and AGS cell lines using Sanger sequencing method to make sure these cell lines used in our research were rs13042395 CC genotype.
Electrophoresis mobility shift assay (EMSA) analysis was performed at Viagene Co. Ltd (Shanghai, China) and was used to detect the different binding affinities of nucleoprotein with the rs13042395 C or T allele flanked by 58bp DNA up‐ and downstream. The DNA sequence of rs13042395 C allele flanked by 58 bp nucleotide was as follows: 5′‐TGGGGTTCTGACCAGGGCCAGTGCACCGTCATTGTGTGGGCTGGGCCATCTCCTCCAGG‐3’. The rs13042395 T allele flanked by 58 bp nucleotide was as follows: 5’‐TGGGGTTCTGACCAGGGCCAGTGCACCGTTATTGTGTGGGCTGGGCCATCTCCTCCAGG‐3’. Bold characters in the sequence represent the rs13042395 locus. The probe representing the rs13042395 C allele (bio‐CLwild) was as follow: 5’‐CCAGTGCACCGTCATTGTGTGGGCT‐3’, and the probe representing the rs13042395 T allele (bio‐CLmut) was as follow: 5’‐CCAGTGCACCGTTATTGTGTGGGCT‐3’. The positive control probe was as follow: 5’‐CCAGTGCAAAGAGCTTGTGTGGGCT‐3’, and the negative control probe was as follow: 5’‐CGTACGCGCTGTCATATTGACAGGT‐3’. The nucleoprotein of BGC823 and AGS cell lines were extracted and incubated with different probes which were labelled with biotin. After that, nucleoprotein‐probes complex were separated by electrophoresis and then biotin signals were detected to assess the quantities of nucleoprotein‐probes complex.
In the competitive EMSA assay, the probe representing the rs13042395 C allele, which was not labelled with biotin was called CLwild. The standard probe binding MEIS1 protein (std‐ME1S1) was as follow: 5’‐CCAGTGCACTGTCATTGTGTGGGCT‐3’. The probe with modifications within the key motif binding MEIS1 (pc‐MEIS1) was as follow: 5’‐ CCAGTGCAAAGAGCTTGTGTGGGCT ‐3’. The probe with modifications outside the key motif binding MEIS1 (nc‐MEIS1) was as follow: 5’‐ CGTACGCGCTGTCATATTGACAGGT ‐3’. All the four above probes were not labelled with biotin and were, respectively, added into bio‐CLwild probes and BGC823 cell nucleoprotein to competitively bind to the transcription factors, assumedly the MEIS1.
In the super‐shift EMSA assay, MEIS1 antibodies were added to ascertain whether the DNA‐protein complex can bind the antibodies.
2.5. Cell lines and culture
Human gastric cancer lines BGC823, MGC803 and AGS were obtained from the Cell Bank of Type Culture Collection of Chinese Academy of Sciences (Shanghai, China). All of the above cell lines have been authenticated by short tandem repeat (STR) DNA profiling analysis. Cells were cultured in RPMI 1640 medium supplemented with 10% FBS (Gibco, USA) and antibiotics at 37°C, 5% CO2.
2.6. Gene sets enrichment analysis, lentiviruses and infection
The gene sets enrichment analysis (GSEA) of two Gene Expression Omnibus (GEO) data sets (GSE62254 and GSE15459) and TCGA gastric cancer data set were conducted to calculated the most likely downstream effector gene of SLC52A3.
Lentiviral vectors expressing SLC52A3 and Gap junction protein alpha 1 (GJA1) were obtained from Shanghai Hanyin Biotechnology Co Ltd. MGC803 and AGS cells were infected with lentivirus carrying SLC52A3 or GJA1, and transfected cells were selected with indicated antibiotics to generate overexpressing stable cells. Western blotting was applied to detect the expression level of SLC52A3 and GJA1, and GAPDH was used as the internal reference protein.
2.7. Western blotting and PCR
Firstly, the whole cell lysates were obtained as the supernatant, and protein concentration was determined using the Pierce BCA Assay Kit (Thermo Scientific). Then, a total of 30 μg protein was loaded into each lane and separated by a 10% SDS‐PAGE gel. After that, proteins were transferred into PVDF membranes. The membranes were blocked with 5% of skim milk for one hour at room temperature and incubated in primary antibodies at 4°C overnight. The next day, the membranes were washed with TBST and incubated with HRP‐conjugated secondary antibody for one hour at room temperature. Finally, the protein bands were detected with enhanced chemiluminescence (Millipore, Burlington, MA, USA) by a luminescent image analyser (ImageQuant LAS4000). Primary antibodies against MEIS1 were purchased from Active Motif (Carlsbad, California, USA), SLC52A3 and GJA1 were purchased from Abcam (Cambridge, UK), primary antibody against GAPDH and the secondary antibodies were from Proteintech (Wuhan, China).
RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). A PrimeScript reagent Kit with gDNA Eraser (Takara, Dalian, China) was used to reverse transcribe RNA into cDNA, and then, quantitative real‐time PCR was performed with TB Green Ex TaqTM (Takara) using Applied Biosystems Prism 7900 system (Life Technologies, Carlsbad, CA, USA). The sequences of the primers were as follows: MEIS1:5’‐CTTCCCTCTCTTAGCACTGATT‐3’; SLC52A3: 5’‐GCATCGCCTTCTTGGTCCTCAC‐3’; GAPDH: 5’‐ACCCAGAAGACTGTGGATGG‐3’.
2.8. Proliferation, migration and invasion assay
The EdU cell proliferation assay and plate colony formation assay were used to evaluate the ability of cell proliferation. For EdU cell proliferation assay, cells were seeded into 24‐well plates with a density of 5 × 104 cells per well. After 24‐hour culture, cells were incubated with EdU for 4 hours, then fixed with 4% paraformaldehyde and permeabilized by 0.5% Triton X‐100. After that, EdU staining and nuclear staining with DAPI were applied to identify proliferative cells. Finally, images were captured using confocal microscopy and the proportion of EdU‐positive cells was calculated. For plate colony formation assay, cells were seeded into 6‐well plates with a density of 1 × 103 per well. After 10‐day culture, colonies were fixed with 4% paraformaldehyde, stained with 0.1% crystal violet and counted. All experiments were performed in triplicate. The transwell assay was used to assess cell migration and invasion with the transwell system (24‐well insert, pore size, 8 μm; Corning co. Ltd, Corning, New York, USA) For migration assay, 41 × 104 cells in 200 μL serum‐free medium were plated into the upper chamber, and 600 μL medium containing 20% FBS was added into the lower chamber. 24 hours later, the cells that migrated to the lower side of chamber membrane were fixed with 4% paraformaldehyde, stained with 0.1% crystal violet and counted. For invasion assay, Matrigel (1:10) was polymerized in upper chamber for 45 minutes at 37°C, then 11 × 105 cells in serum‐free medium were plated into the upper chamber, and 600 μL medium containing 20% FBS was added into the lower chamber. 48 hours later, the cells that invaded to the lower side of chamber membrane were fixed with 4% paraformaldehyde, stained with 0.1% crystal violet and counted.
All experiments were performed with mycoplasma‐free cells.
2.9. Statistic analysis
The survival analysis was assessed by KM method, log‐rank test and Cox proportional hazards regression. The expressional differences of SLC52A3 gene were analysed by unpaired t test. The statistical analysis was performed by R language (version 3.5.1). P values were two‐sided with a significance level of 0.05.
3. RESULTS
3.1. Clinicopathological characteristics of the research population
Our research recruited 926 GCa patients, details were included in Table 1. In them, 48% (444) patients were stage I‐II and 52% (482) were stage III‐IV. Over 88% (820) of these patients were pathologically diagnosed adenocarcinoma and with 11.4% (106) of these patients were diagnosed with mucinous adenocarcinoma. 84.2% (784) patients underwent surgery and 15.2% (140) patients did not; 73.2% (678) patients underwent chemotherapy, and 26.8% (248) patients did not receive any chemotherapies.
TABLE 1.
Clinicopathological characteristics of FUSCC GCa patients included in the present study
| Variable | Case No. (100%) |
|---|---|
| All patients | 926 |
| Age (y) | |
| ≤59 | 477 (51.5) |
| >59 | 449 (48.5) |
| Sex | |
| Male | 658 (71.1) |
| Female | 268 (28.9) |
| Smoking | |
| Yes | 565 (61.0) |
| No | 361 (39.0) |
| Alcohol | |
| Yes | 218 (23.5) |
| No | 708 (76.5) |
| BMI | |
| <23 | 538 (58.1) |
| ≥23 | 388 (41.9) |
| Chemotherapy | |
| Yes | 678 (73.2) |
| No | 248 (26.8) |
| Surgery | |
| Yes | 784 (84.8) |
| No | 140 (15.2) |
| Stage | |
| I‐II | 444 (48.0) |
| III‐IV | 482 (52.0) |
| Pathological type | |
| Adenocarcinoma | 820 (88.6) |
| Mucus adenocarcinoma/SRCC | 106 (11.4) |
| Differentiation grade | |
| Well‐moderate | 214 (23.1) |
| Poor | 712 (76.9) |
Abbreviations: BMI, body mass index; FUSCC, Fudan University Shanghai Cancer Center; GCa, gastric carcinoma; SRCC, signet‐ring cell carcinoma.
3.2. SLC52A3 rs13042395 C allele predict better survival in GCa patients
In Figure 1, we demonstrated that SLC52A3 rs13042395 C > T change was significantly associated with OS in GCa patients with recessive model (ie TC/CC genotype versus TT genotype, log‐rankP = 0.015). After adjusted by clinical variables, SLC52A3 rs13042395 was proved to be an independent prognosis factor by multivariate Cox analysis (HR = 0.57, 95%CI (0.40‐0.82), and P = 0.031). In the stratification analysis, SLC52A3 rs13042395 TC/CC genotype favoured the better survival in all subgroups except the signet‐ring cell carcinoma GCa subgroup.
FIGURE 1.
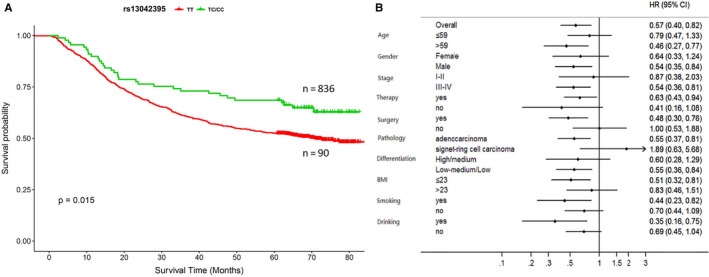
SLC52A3 rs13042395 predicted survival of GCa patients and stratification analysis. (A) Patients of TC/CC genotype (n = 836) had a significant longer survival than the patients of TT genotype (n = 90). (B) TC/CC genotype favoured longer survival in all subgroups except the signet‐ring cell carcinoma subgroups. GCa, gastric carcinoma; BMI, body mass index
3.3. SLC52A3 rs13042395 T allele significantly increase the SLC52A3 expression by altering its binding affinities with MEIS1 compared to rs13042395 C allele
From GTEx database, we found that rs13042395 C > T change was significantly associated with higher SLC52A3 mRNA expression level in normal gastric tissue (P = 0.0029) (Figure 2A).
FIGURE 2.
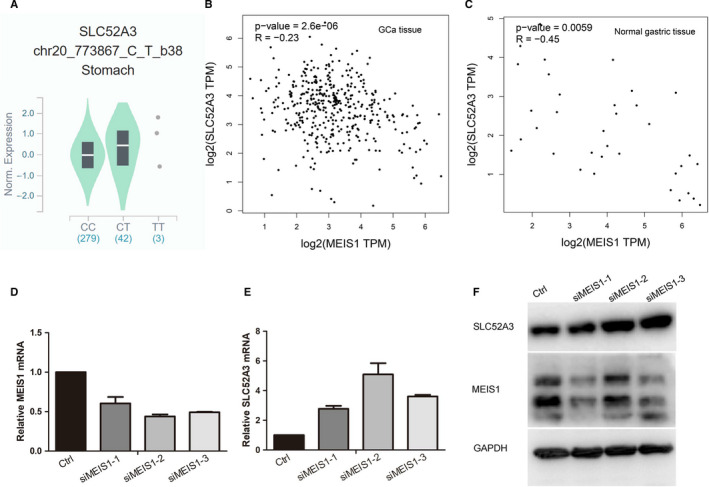
SLC52A3 rs13042395 C > T change down‐regulated its mRNA expression via changing binding affinity with MEIS1. (A) In normal gastric tissue, samples with rs13042395 CT genotype had significantly higher expression level of SLC52A3 compared to CC genotype based on GTEx database. (B‐C) In GCa tissues and normal gastric tissues, the expression of SLC52A3 was negatively significantly associated with the MEIS1 gene from GEPIA online tools based on TCGA database. (D‐F) Knock‐down MEIS1 in BGC823 cells using three different siRNA led to the decreased expression of SLC52A3 mRNA and protein. GCa, gastric carcinoma; GTEx, The Genotype‐Tissue Expression project; TCGA, The Cancer Genome Atlas
According to the Jaspar web server, among the transcription factors which could bind to the SLC52A3 rs13042395, the transcription factor MEIS1 was found to have the biggest binding affinity change (specifically, the binding affinity of MEIS1 decreased) when SLC52A3 rs13042395 C > T change occurred (Table S1). The MEIS1 expression was negatively associated with SLC52A3 gene expression in GCa tissue or normal gastric tissue according to the GEPIA web server which was based on TCGA data sets (Pierson correlation, R = −0.23,P < 0.00001 and R = −0.45, P < 0.01, respectively) (Figure 2B‐C). After knock‐down, the MEIS1 using siRNA in the BGC823 cells, the SLC52A3 mRNA and protein expression decreased significantly (Figure 2D‐F), which validated that MEIS1 played as an inhibitory transcription factor in regulating the SLC52A3 gene.
Targeting sequencing showed that MGC803, AGS and BGC823 cell all carried rs13042395 CC genotype (Figure S1A). In EMSA analysis, it was revealed that the binding affinity of rs13042395 T allele probes with nucleoprotein was weaker than the rs13042395 C allele in BGC 823 cell line while this interaction of probes and nucleoprotein was not observed in AGS cell line (Figure 3A). In the competitive EMSA assay (Figure 3B), binding affinity of rs13042395 C allele probes (ie bio‐CLwild probe) with MGC823 cell nucleoprotein was attenuated after adding the rs13042395 C allele probes without labelled with biotin (ie CLwild probe), and this effect was amplified when the concentration was elevated from 40 µmol/L to 80 µmol/L. The binding affinity of bio‐CLwild probe with nucleoprotein was also attenuated by adding the standard probes for detecting the MEIS1 protein (ie std‐MEIS1 probe), and also, this effect was amplified when the concentration was elevated from 40 µmol/L to 80 µmol/L. To ensure that it was the MEIS1 transcription factor which bound to bio‐CLwild probes, we further designed the probes with modifications within the motif binding MEIS1 (ie pc‐MEIS1) and probes with modifications outside the motif binding MEIS1 (ie nc‐MEIS1). It was showed that the nc‐MEIS1 probes attenuated the binding affinity of bio‐CLwild probe with BGC823 cell nucleoprotein, and this attenuating effect was amplified when the concentration was elevated from 40 µmol/L to 80 µmol/L. However, the pc‐MEIS1 probes showed no effect on the binding affinity of bio‐CLwild probe with BGC823 cell nucleoprotein whether the concentration was 40 µmol/L or 80 µmol/L. In the super‐shift EMSA assay (Figure 3C), a super‐shifted band was demonstrated after adding MEIS1 antibody.
FIGURE 3.
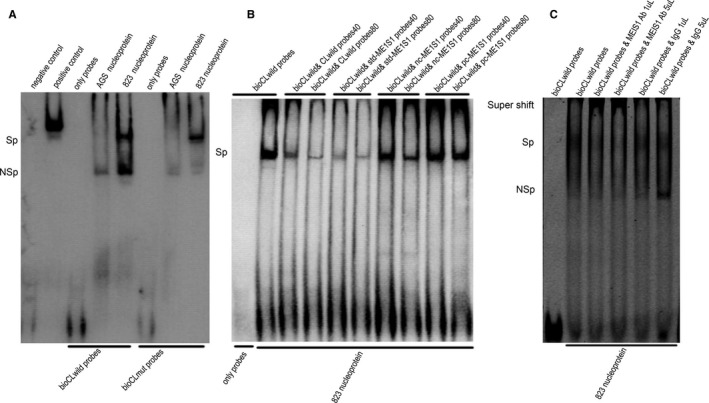
SLC52A3 rs13042395 C allele had more firm binding affinity with MEIS1 than the T allele. (A) SLC52A3 rs13042395 C allele had higher binding affinities to nucleoprotein compared to T allele in BGC823 cells. (B) competitive EMSA revealed that SLC52A3 rs13042395 C allele had higher binding affinities to MEIS1 compared to T allele. (C) super‐shift EMSA revealed that MEIS1 antibodies could bind to DNA‐protein complex leading to a super‐shift band. GCa, gastric carcinoma; GTEx, The Genotype‐Tissue Expression project; TCGA, The Cancer Genome Atlas; bio‐CLwild, the probe representing the rs13042395 C allele and labelled with biotin; CLwild, the probe representing the rs13042395 C allele and without labelled with biotin; bio‐CLmut, The probe representing the rs13042395 T allele and labelled with biotin; std‐MEIS1, The standard probe binding MEIS1 protein; pc‐MEIS1, The probe with modifications within the motif binding MEIS1 protein; nc‐MEIS1, the probe with modifications outside the motif binding MEIS1 protein; 40 represents 40 µmol/L; 80 represents 80 µmol/L; Ab, antibody; Sp, specific binding; NSp, non‐specific binding
Based on the above assays, it was found that the SLC52A3 rs13042395 C allele could more firmly bind to the MEIS1 transcription factor than the T allele, and the MEIS1 gene served as an inhibitory transcription in regulating the SLC52A3, so that SLC52A3 rs13042395 C > T change resulted in increased expression level of SLC52A3.
3.4. SLC52A3 positively regulates malignant phenotype of gastric cancer cells
To explore the role of SLC52A3 in regulating malignant phenotype of gastric cancer cells, including cell proliferation, colony formation, migration and invasion, we established stable transfected gastric cancer cell lines with MGC803 and AGS. Western blotting verified successful overexpression of SLC52A3 in MGC803 and AGS as shown in (Figure 4D). The results of EdU cell proliferation assay and colony formation assay showed that overexpression of SLC52A3 in both MGC803 and AGS increased rate of EdU‐positive cells and number of clones, which indicated that SLC52A3 could promote gastric cancer cells proliferation (Figure 4A‐C, E‐F). Our results of transwell chamber assay demonstrated that overexpression of SLC52A3 could promote gastric cancer cells migration and invasion in MGC803 and AGS (Figure 4G‐J).
FIGURE 4.
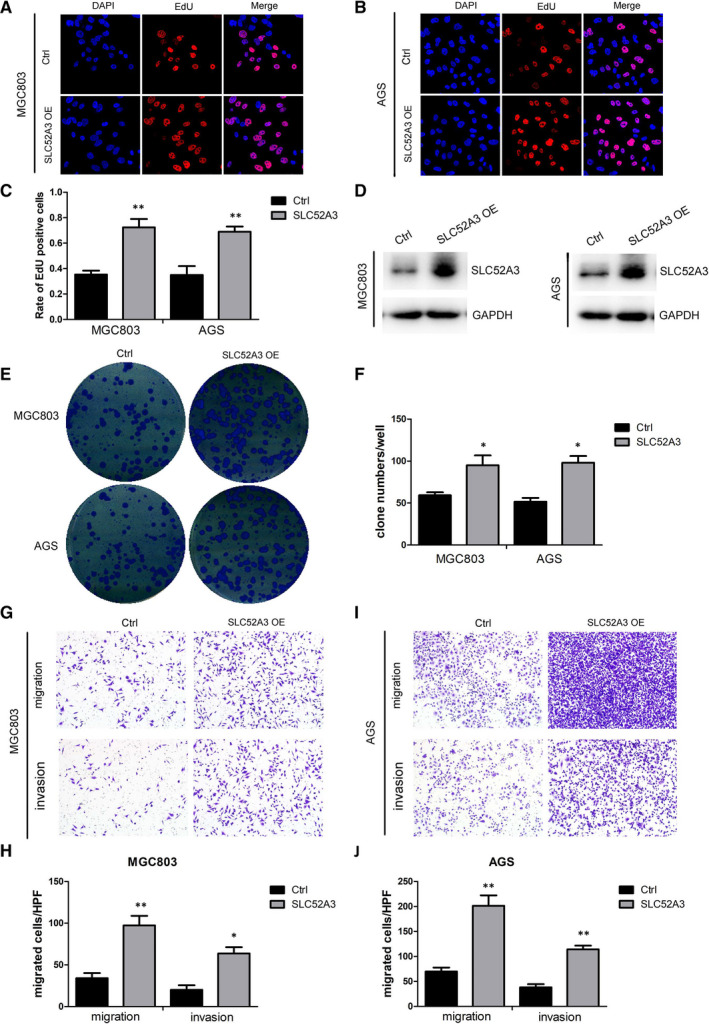
Overexpression of SLC52A3 promoted malignant phenotype of GCa cells. SLC52A3 was overexpressed using lentiviral vectors in MGC803 cells and AGS cells, and western blotting was used to verify the overexpression of SLC52A3 (D). The aggressive performance was assessed by EdU cell proliferation assay (A: MGC803,B: AGS, C: quantification of A and B), colony formation (E‐F), and transwell assay (G: MGC803, I: AGS, H‐J: quantification of G and I). GCa, gastric carcinoma. *P < 0.05 **P < 0.01
3.5. SLC52A3 regulates malignant phenotype of gastric cancer cells through down‐regulation of GJA1
The GSEA of two GEO data sets (GSE62254 and GSE15459) and TCGA gastric cancer data set revealed that GJA1 may be a downstream target gene for SLC52A3 (Figure S2), we further investigated whether SLC52A3 regulated malignant phenotype of gastric cancer cells via regulating GJA1. We firstly detected GJA1 expression in SLC52A3 overexpression gastric cancer cells and found that overexpression of SLC52A3 resulted in decreased GJA1 (Figure 5A). To further explore the role of GJA1 in SLC52A3‐mediated malignant phenotype, we co‐transfected GJA1 in SLC52A3‐expressing MGC803 and AGS gastric cancer cells and detected GJA1 expression level by Western blotting (Figure 5B). Our results showed that overexpression of SLC52A3 promoted cell proliferation, colony formation, migration and invasion, while GJA1 overexpression inhibited malignant phenotype (Figure 5C‐G). Furthermore, co‐transfection of SLC52A3 and GJA1 restored proliferation, colony formation, migration and invasion of gastric cancer cells (Figure 5C‐G). The above results demonstrated that SLC52A3 regulated malignant phenotype of gastric cancer cells through down‐regulation of GJA1. Additionally, Knockdown the MEIS1 in BGC823 cells led to the decreased expression of GJA1 protein (Figure S1B).
FIGURE 5.
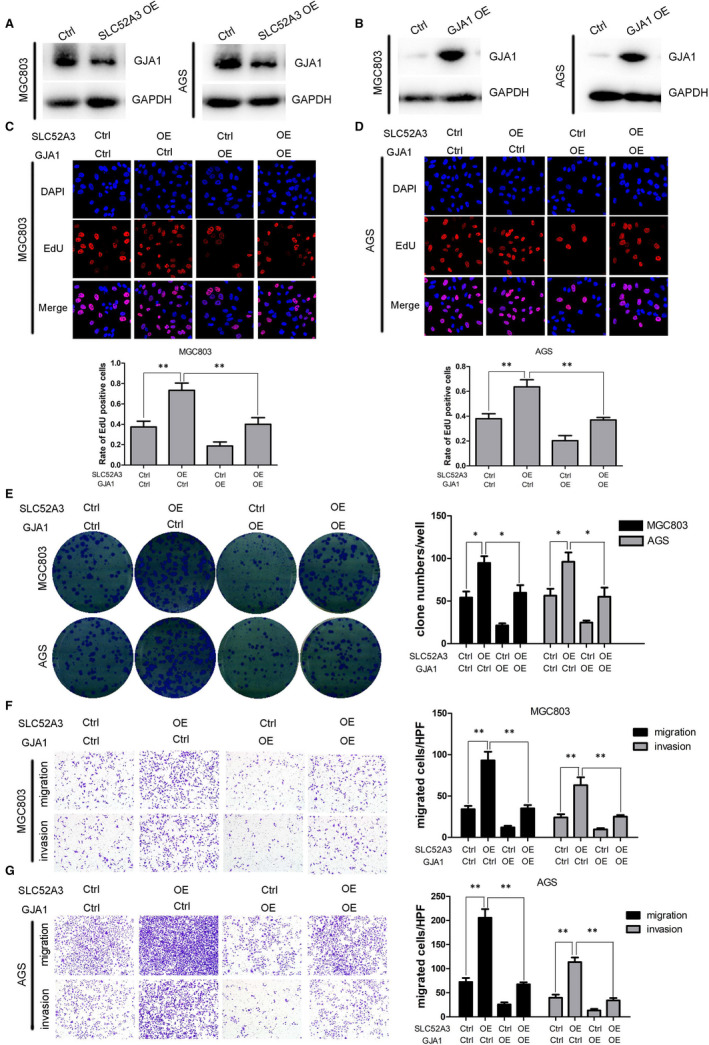
The interaction of SLC52A3 and GJA1 in GCa cells. Overexpression SLC52A3 down‐regulated GJA1 expression (A), GJA1 overexpression by lentiviral vectors was validated by Western blotting assay (B). Overexpression of SLC52A3 promoted cell proliferation, colony formation, migration and invasion, while GJA1 overexpression inhibited malignant phenotype (C‐G). Co‐transfection of SLC52A3 and GJA1 restored proliferation, colony formation, migration and invasion of GCa cells (C‐G). GCa, gastric carcinoma. *P < 0.05, **P < 0.05
4. DISCUSSION
It was well established that precise prognosis prediction played an important role for treatment decision‐making for GCa, for the reason that high‐risk patients may need more aggressive interventions and low‐risk patients need avoid overtreatments. 11 In this relatively large survival analysis in Chinese GCa patients which contained over 900 patients, we identified that SLC52A3 rs13042395 C > T variation would independently significantly impact the OS of patients and demonstrated that SLC52A3 expression was increased due to the different binding affinities of rs13042395 C > T change with transcriptional factors. Specifically, rs13042395 C allele had a higher binding affinity with inhibitory transcription factors MEIS1 compared to the rs13042395 T allele so that rs13042395 T allele significantly increased the expression of SLC52A3 compared to C allele. Further mechanism investigation found that overexpression of SLC52A3 contributes to the increased ability of proliferation, colony formation, migration and invasion likely via down‐regulation of GJA1 expression in vitro.
SLC52A3 was essential for absorption of riboflavin, which was a critical component of the mitochondrial electron transport chain, and loss functional mutation of SLC52A3 may lead to Brown‐Vialetto‐Van Laere syndrome, a rare neurological disorder characterized by bulbar palsies and sensorineural deafness. 12 As for the cancer risk, riboflavin supplement was thought to reduce various types of cancer risks, 13 and several cancer risk research found that SLC52A3 SNPs was associated with oesophageal cancer risk, 14 , 15 , 16 probably activated by NF‐κB p65/Rel‐B. 17 Previous studies also found that SLC52A3 rs13042395 C > T change could promote glioma cancer cell progression and migration in vivo and in vitro. 18 Interestingly, researchers found that SLC52A3 rs13042395 TT genotype favoured reduced lymph node metastasis rate and longer relapse‐free survival in oesophageal squamous cell carcinoma. 19 Meanwhile, other researcher found that in oesophageal cancer cells, depletion of SLC52A3 would increase p21 and p27 protein levels, decreased cyclin E1 and Cdk2 leading to cell cycle arrest at G1‐G1/S, 8 suggested that SLC52A3 rs13042395 C > T change played a complicated role in oesophageal squamous cell carcinoma. While in the GCa, as 90% of them were adenocarcinoma, SLC52A3 rs13042395 C > T change played a consistent role as an oncogene both in risk or survival analysis and in our present mechanism studies.
The downstream target genes of SLC52A3 were unclear in GCa cells so far as we know. Using GSEA analysis, we speculated that the GJA1 may be a target gene of SLC52A3 in GCa cells. Further research in vitro confirmed that GJA1 gene was negative regulated by SLC52A3 gene in our study.
GJA1, which was enriched in the downstream of SLC52A3 according to our research, was a member of the connexin gene family and encoded cell gap junction protein, and it was crucial for low molecular weight materials transportation from the cell to cell. 20 Researchers already confirmed that the connexin gene family played a complicated role in tumorigenesis and progression. 21 Recently researcher found that GJA1 was associated in glioblastoma cancer cells apoptosis and promote the progression and invasion in breast cancer cells. 22 , 23 Few research about GJA1 was conducted in GCa, one of them suggested that overexpression of GJA1 leads to decreased ability of colony forming and invasive ability in BGC‐823 cells, 24 which was consistent with our research results.
In conclusion, we found that SLC52A3 rs13042395 C > T change independently predicted the survival in Chines eastern GCa patients. SLC52A3 rs13042395 T allele had a lower binding affinity with inhibitory transcription factor, MEIS1, leading the up‐regulation of SLC52A3 gene, SLC52A3 overexpression is associated with aggressive phenotype in GCa cells likely via down‐regulation of GJA1 gene. However, because our research was conducted just in one medical centre and only include Han ethnicity patients, additional multi‐centres and multi‐ethnicities studies or clinical trials as well as mechanism investigations in vivo are needed to confirm these results.
CONFLICTS OF INTEREST
The authors have no conflicts of interest to declare.
AUTHOR CONTRIBUTION
Xiaofei Qu: Conceptualization (lead); Data curation (equal); Formal analysis (equal); Investigation (equal); Methodology (equal); Software (lead); Writing‐original draft (equal). Lei Cheng: Conceptualization (equal); Data curation (equal); Formal analysis (equal); Funding acquisition (supporting); Investigation (equal); Methodology (equal); Writing‐review & editing (equal). Liqin Zhao: Formal analysis (equal); Investigation (equal); Project administration (equal); Writing‐original draft (equal). Lixin Qiu: Conceptualization (supporting); Data curation (supporting); Methodology (equal); Project administration (lead); Supervision (equal); Validation (equal); Writing‐review & editing (lead). Wejian Guo: Conceptualization (equal); Funding acquisition (lead); Project administration (lead); Resources (lead); Supervision (lead); Writing‐original draft (supporting); Writing‐review & editing (lead).
Supporting information
Fig S1
Fig S2
Table S1
ACKNOWLEDGEMENTS
We gratefully acknowledge the financial supports by the National Science Foundation of China, and we also gratefully acknowledge the tissue bank of Fudan University Shanghai Cancer Center (FUSCC) for their assistance and kindly advise. This work was supported by the Natural Science Foundation of China (grant numbers. 81101808 and 81902875).
Qu X, Cheng L, Zhao L, Qiu L, Guo W. Functional variation of SLC52A3 rs13042395 predicts survival of Chinese gastric cancer patients. J Cell Mol Med. 2020;24:12550–12559. 10.1111/jcmm.15798
Xiaofei Qu, Lei Cheng and Liqin Zhao are contributed equally to this work.
Contributor Information
Lixin Qiu, Email: mdqiulixin@hotmail.com.
Weijian Guo, Email: guoweijian_fudan@163.com.
DATA AVAILABILITY STATEMENT
The deidentification data that support the findings of this study are available on request from the corresponding author. The raw data are not publicly available due to privacy or ethical restrictions, any researcher who are interested in getting raw data are welcome to contact Fudan University institutional review board to obtain permission.
REFERENCES
- 1. Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA: Cancer J Clinic. 2016;66(2):115‐132. [DOI] [PubMed] [Google Scholar]
- 2. Digklia A, Wagner AD. Advanced gastric cancer: Current treatment landscape and future perspectives. World J Gastroenterol. 2016;22(8):2403‐2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dragomir MP, Kopetz S, Ajani JA, Calin GA. Non‐coding RNAs in GI cancers: from cancer hallmarks to clinical utility. Gut. 2020;69(4):748‐763. [DOI] [PubMed] [Google Scholar]
- 4. Hu J, Yu J, Gan J, et al. Notch1/2/3/4 are prognostic biomarker and correlated with immune infiltrates in gastric cancer. Aging. 2020;12(3):2595‐2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang W, Li Z, Wang J, et al. A functional polymorphism in TFF1 promoter is associated with the risk and prognosis of gastric cancer. Int J Cancer. 2018;142(9):1805‐1816. [DOI] [PubMed] [Google Scholar]
- 6. Jia ZF, Cao DH, Wu YH, et al. Lethal‐7‐related polymorphisms are associated with susceptibility to and prognosis of gastric cancer. World J Gastroenterol. 2019;25(8):1012‐1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fujimura M, Yamamoto S, Murata T, et al. Functional characteristics of the human ortholog of riboflavin transporter 2 and riboflavin‐responsive expression of its rat ortholog in the small intestine indicate its involvement in riboflavin absorption. J Nutr. 2010;140(10):1722‐1727. [DOI] [PubMed] [Google Scholar]
- 8. Jiang XR, Yu XY, Fan JH, et al. RFT2 is overexpressed in esophageal squamous cell carcinoma and promotes tumorigenesis by sustaining cell proliferation and protecting against cell death. Cancer Lett. 2014;353(1):78‐86. [DOI] [PubMed] [Google Scholar]
- 9. Shi Y, Hu Z, Wu C, et al. A genome‐wide association study identifies new susceptibility loci for non‐cardia gastric cancer at 3q13.31 and 5p13.1. Nat Genet. 2011;43(12):1215‐1218. [DOI] [PubMed] [Google Scholar]
- 10. Mocellin S, Verdi D, Pooley KA, Nitti D. Genetic variation and gastric cancer risk: a field synopsis and meta‐analysis. Gut. 2015;64(8):1209‐1219. [DOI] [PubMed] [Google Scholar]
- 11. Jiang Y, Li T, Liang X, et al. Association of Adjuvant Chemotherapy With Survival in Patients With Stage II or III Gastric Cancer. JAMA Surg. 2017;152(7):e171087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Manole A, Jaunmuktane Z, Hargreaves I, et al. Clinical, pathological and functional characterization of riboflavin‐responsive neuropathy. Brain. 2017;140(11):2820‐2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang SM, Taylor PR, Fan JH, et al. Effects of Nutrition Intervention on Total and Cancer Mortality: 25‐Year Post‐trial Follow‐up of the 5.25‐Year Linxian Nutrition Intervention Trial. J Natl Cancer Inst. 2018;110(11):1229‐1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gu H, Ding G, Zhang W, et al. Replication study of PLCE1 and C20orf54 polymorphism and risk of esophageal cancer in a Chinese population. Mol Biol Rep. 2012;39(9):9105‐9111. [DOI] [PubMed] [Google Scholar]
- 15. Wei W, Ji A, Wang J, et al. Functional single nucleotide polymorphism in C20orf54 modifies susceptibility to esophageal squamous cell carcinoma. Dis Esophagus. 2013;26(1):97‐103. [DOI] [PubMed] [Google Scholar]
- 16. Li J, Wang S, Li M, et al. Decreased risk of developing cancer in subjects carrying SLC52A3 rs13042395 polymorphism: proof from a meta‐analysis. Biomark Med. 2016;10(10):1105‐1118. [DOI] [PubMed] [Google Scholar]
- 17. Long L, Pang XX, Lei F, et al. SLC52A3 expression is activated by NF‐kappaB p65/Rel‐B and serves as a prognostic biomarker in esophageal cancer. Cell Mol Life Sci. 2018;75(14):2643‐2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fu T, Liu Y, Wang Q, et al. Overexpression of riboflavin transporter 2 contributes toward progression and invasion of glioma. NeuroReport. 2016;27(15):1167‐1173. [DOI] [PubMed] [Google Scholar]
- 19. Tan HZ, Wu ZY, Wu JY, et al. Single nucleotide polymorphism rs13042395 in the SLC52A3 gene as a biomarker for regional lymph node metastasis and relapse‐free survival of esophageal squamous cell carcinoma patients. BMC Cancer. 2016;16:560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Batra N, Kar R, Jiang JX. Gap junctions and hemichannels in signal transmission, function and development of bone. Biochim Biophys Acta. 2012;1818(8):1909‐1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Aasen T, Mesnil M, Naus CC, Lampe PD, Laird DW. Gap junctions and cancer: communicating for 50 years. Nat Rev Cancer. 2016;16(12):775‐788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Huang BR, Tsai CH, Chen CC, et al. Curcumin promotes Connexin 43 degradation and temozolomide‐induced apoptosis in Glioblastoma cells. Am J Chin Med. 2019;47(3):657‐674. [DOI] [PubMed] [Google Scholar]
- 23. Busby M, Hallett MT, Plante I. The complex subtype‐dependent role of Connexin 43 (GJA1) in breast cancer. Int J Mol Sci. 2018;19(3):693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu D, Zhou H, Wu J, et al. Infection by Cx43 adenovirus increased chemotherapy sensitivity in human gastric cancer BGC‐823 cells: not involving in induction of cell apoptosis. Gene. 2015;574(2):217‐224. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1
Fig S2
Table S1
Data Availability Statement
The deidentification data that support the findings of this study are available on request from the corresponding author. The raw data are not publicly available due to privacy or ethical restrictions, any researcher who are interested in getting raw data are welcome to contact Fudan University institutional review board to obtain permission.


