Fig. 2.
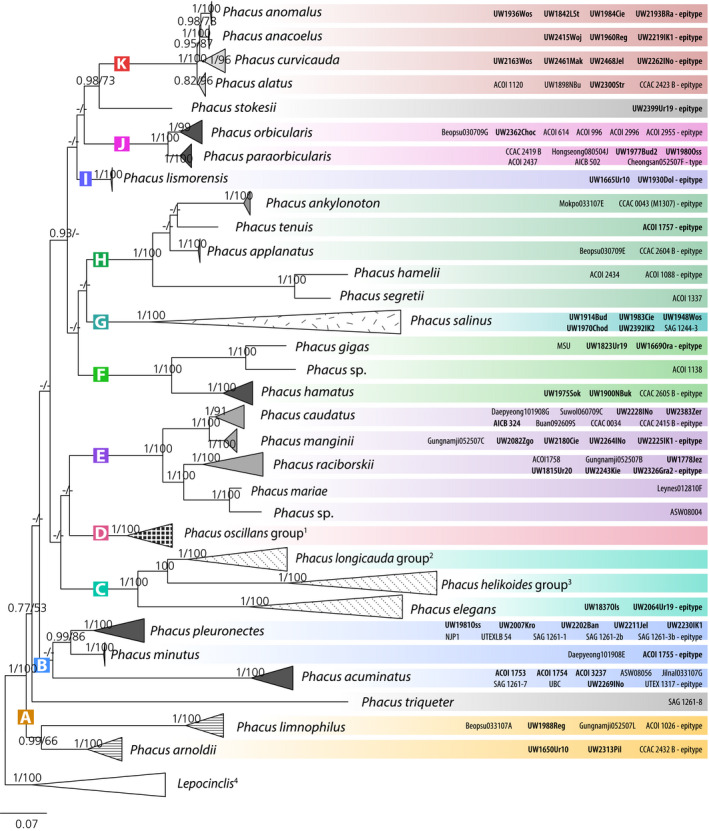
Consensus Maximum Likelihod tree based on 136 nSSU rDNA sequences (of which 129 represent Phacus). Isolates or strains with identical nSSU rDNA are represented by single tips. Sequences of the same species are represented by triangles of height proportional to the variability of sequences. Nodes are labeled with the Bayesian posterior probability (pp) values and rapid bootstrap (rbs) values. The pp values <0.75 and rbs values <50 and clades not present in the particular analysis are marked with a hyphen (‐). Scale bar represents number of substitutions per site. Sequences obtained in this study are indicated in bold type.
Groups of species:
1 P. brevisulcus Suwol060709A (type), P. oscillans ACOI 1339 (epitype), P. longisulcus Psurononuma100609J (type), P. smulkowskianus ACOI 1226 (epitype), UW2281Kop, P. granum AICB349, P. polytrophos CCAC2451B (epitype), P. minimus Buan092609I (type), P. claviformis Gungnamji052507F (type), P. hordeiformis Yongho092609A (type), P. inflexus ACOI 1336 (epitype), P. parvulus ACOI 1093 (epitype), P. viridioryza Sondang060709L (type), P. pusillus UTEX 1282 (epitype), P. skujae ACOI 1312 (epitype).
2 P. ranula Jigok090112, P. convexus UW1821Ora (epitype), P. tortus UW1845Jel (epitype), P. cordatus UW1808Ur17 (epitype), P. longicauda UW1896Laz (epitype), P. sp. Burni081809, P. circumflexus UW1844Jel (epitype).
3 P. crassus UW1566UR15 (type), P. cristatus UW1929Dol (type), P. helikoides UW1658Ur20 (epitype).
4 L. steinii Cheongsan052507K5, L. hispidula MSU, L. fusiformis ACOI 1025, L. spirogyroides ACOI 1027, L. acus ASW 08037, L. tripteris UWOB, L. fusca Saeraewool102007P. [Color figure can be viewed at wileyonlinelibrary.com]
