Fig. 4.
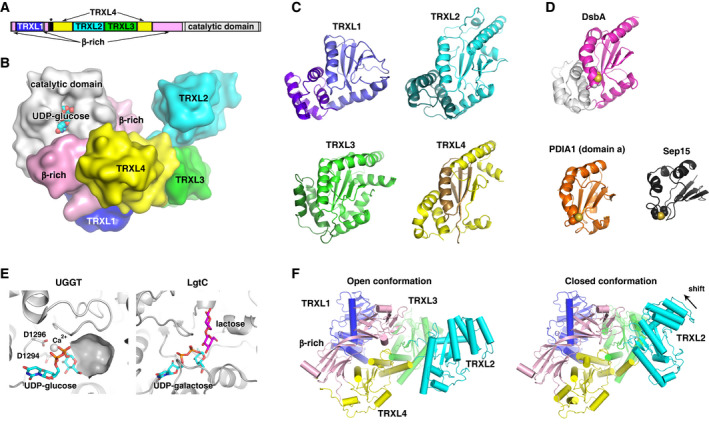
Structure of UDP‐glucose:glycoprotein glucosyltransferase. (A) Domain architecture of UGGT. The Sep15‐binding site is marked with an asterisk. (B) Cartoon representation of UGGT full‐length structure from Chaetomium thermophilum (PDB 5MZO) shows four DsbA‐like domains TRXL1 (blue), TRXL2 (cyan), TRXL3 (green), and TRXL4 (yellow) followed by β‐rich domain (pink) and catalytic domain (gray). Location of UDP‐glucose is modeled from the structure of the Thermomyces dupontii UGGT catalytic domain with bound UDP‐glucose (PDB 5H18). DsbA‐like domains of UGGT (C) have stronger resemblance to bacterial oxidoreductase DsbA (PDB 5KBC; magenta/gray) (D) than to thioredoxin‐like domains in mammalian PDI (PDB 3F8U; orange). Surprisingly, the helical bundle (purple) is at the N terminus of TRXL1 domain, while the TRXL4 domain is assembled from nonconsecutive stretches (yellow and brown) of the primary sequence. The C‐terminal domain of Sep15 (PDB 2A4H; dark gray) represents a minimalist version of a redox fold. (E) The UGGT catalytic domain is similar to other glycosyltransferases. The structure of UGGT catalytic domain (PDB 5H18) with bound UDP‐glucose (cyan) suggests that the acceptor glycan in UGGT binds in a surface cavity formed by a distorted helix next to the glucose moiety of UDP‐glucose. In the structure of the GT8 glycosyltransferases LgtC (PDB 1GA8), this pocket is occupied by the acceptor lactose (purple). Two aspartates in the UGGT catalytic site coordinate a calcium ion. (F) Interdomain mobility of UGGT. Comparison of the open (PDB 5MZO) and closed (PDB 5N2J) UGGT structures of Ch. thermophilum UGGT shows significant differences in positions of the TRXL2 and TRXL3 domains. The catalytic domain has been omitted to better show the conformational changes.
