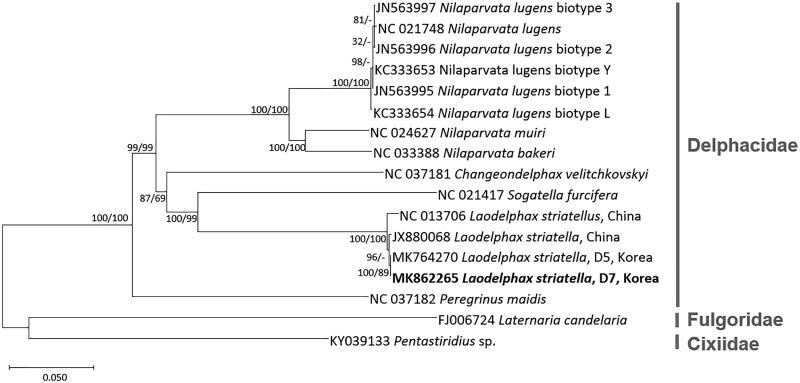
Figure 1.
Neighbour-joining (10,000 bootstrap repeats) and maximum-likelihood (1,000 bootstrap repeats) phylogenetic trees of seven insect species in Delphacidae: four L. striatellus (MK862265 in this study, MK764270, NC_013706, and JX880068), six Nilaparvata lugens (NC_021748, JN563997, JN563996, JN563995, KC333653, and KC333654), Nilaparvata bakeri (NC_033388), Nilaparvata muiri (NC_024627), Sogatella furicefera (NC_021417), Changeondelphax velitchkovskyi (NC_037181), and Peregrinus maidis (NC_037182). As outgroup species, Pentastiridius sp. (KY039133) in Cixiidae and Laternaria candelaria (FJ006724) in Fulgoridae were used. Phylogenetic tree was drawn based on neighbour-joining tree. The numbers above branches indicate bootstrap support values of neighbour-joining and maximum-likelihood phylogenetic trees, respectively.