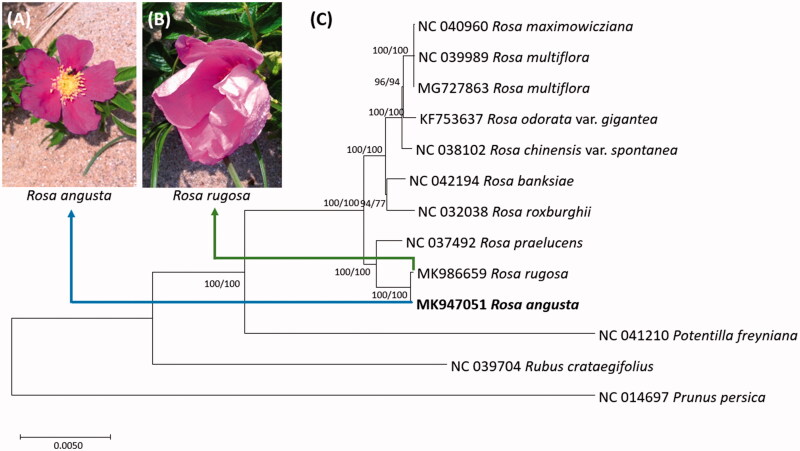
Figure 1.
(A) Picture of Rosa augsta flower, (B) Picture of Rosa rugosa flower, (C) Neighbor joining (bootstrap repeat is 10,000) and maximum likelihood (bootstrap repeat is 1,000) phylogenetic trees of ten Rosa chloroplast genomes and three outgroup species: Rosa angusta (MK947051 in this study), Rosa rugosa (MK986659), Rosa praelucens (NC_037492), Rosa roxburghii (NC_032038), Rosa banksiae (NC_042194), Rosa chinensis var. spontanea (NC_038102), Rosa odorata var. gigantea (KF753637), Rosa multiflora (NC_039989 and MG727863), Rosa maximowicziana (NC_040960), and three outgroup species: Potentilla freyniana (NC_041210), Rubus crataegifolius (NC_039704), and Prunus persica (NC_014697). Phylogenetic tree was drawn based on neighbor joining tree. The numbers above branches indicate bootstrap support values of maximum likelihood and neighbor joining phylogenetic tree, respectively.