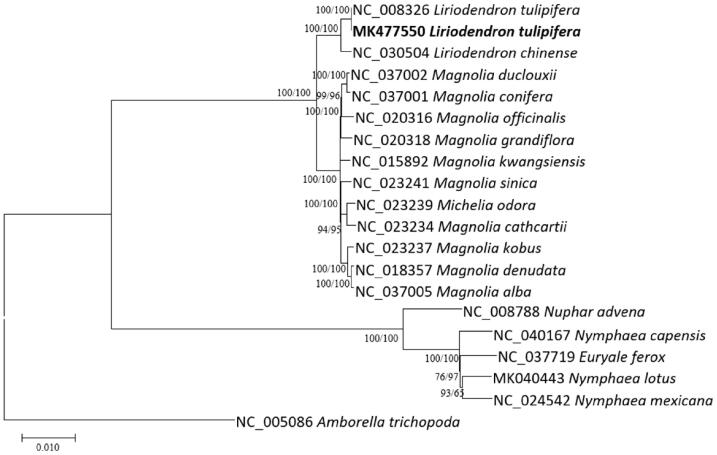
Figure 1.
Neighbor joining (bootstrap repeat is 10,000) and maximum likelihood (bootstrap repeat is 1,000) phylogenetic trees of 14 Magnoliaceae, 4 Nymphaceae, and 1 Amborella complete chloroplast genomes: two Liriodendron tulifipera (MK477550 in this study and NC_008326), Liriodendron chinense (NC_030504), Magnolia alba (NC_037005), Magnolia cathcartii (NC_023234), Magnolia conifera (NC_037001), Magnolia denudata (NC_018357), Magnolia duclouxii (NC_37002), Magnolia grandiflora (NC_020318), Magnolia kobus (NC_023237), Magnolia kwangsiensis (NC_015892), Magnolia officinalis (NC_020316), Magnolia sinica (NC_023241), Michelia odora (NC_023239), Euryale ferox (NC_037719), Nuphar advena (NC_008788), Nymphaea capensis (NC_040167), Nymphaea lotus (MK040443), Nymphaea mexicana (NC_024542), and Amborella trichopoda (NC_005086). The numbers above branches indicate bootstrap support values of maximum likelihood and neighbor joining phylogenetic trees, respectively.