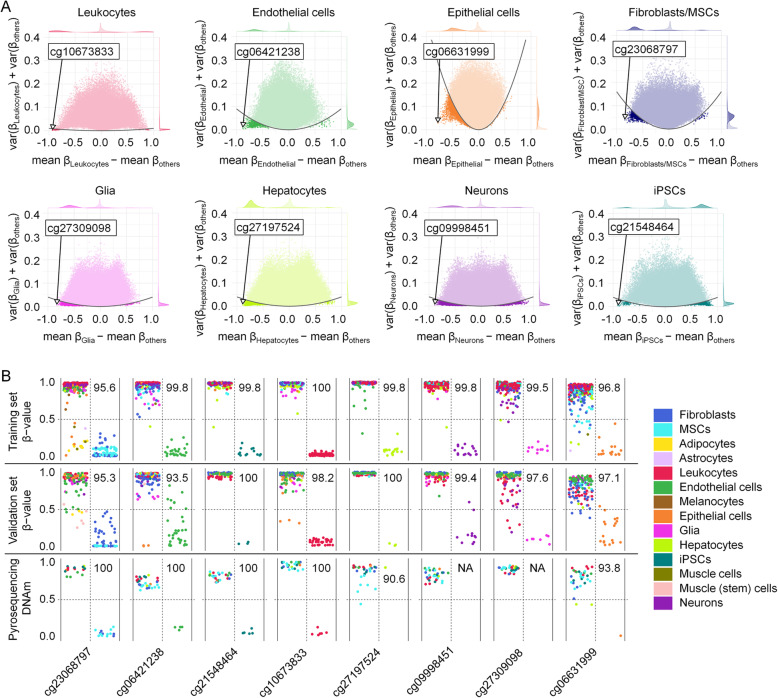
Fig. 3.
Cell type-specific CpG sites are preferentially hypomethylated. a Selection of cell type-specific CpGs for leukocytes, endothelial cells, epithelial cells, fibroblasts/MSCs, glia, hepatocytes, neurons, and iPSCs. The difference of mean β values of each cell type versus all other cell types was plotted against the sum of variances within both groups. CpGs for subsequent deconvolution are highlighted. b DNAm levels of the eight selected CpGs in the training, validation, and pyrosequencing datasets. The vast majority of samples revealed the expected cell type-specific hypomethylation, albeit pyrosequencing of liver cell lines (Hep3B and HuH-7) did not reveal hypomethylation at cg27197524 as expected for primary cells. Glia and neuron samples were not available for pyrosequencing. Numbers correspond to classification accuracy in percentage values