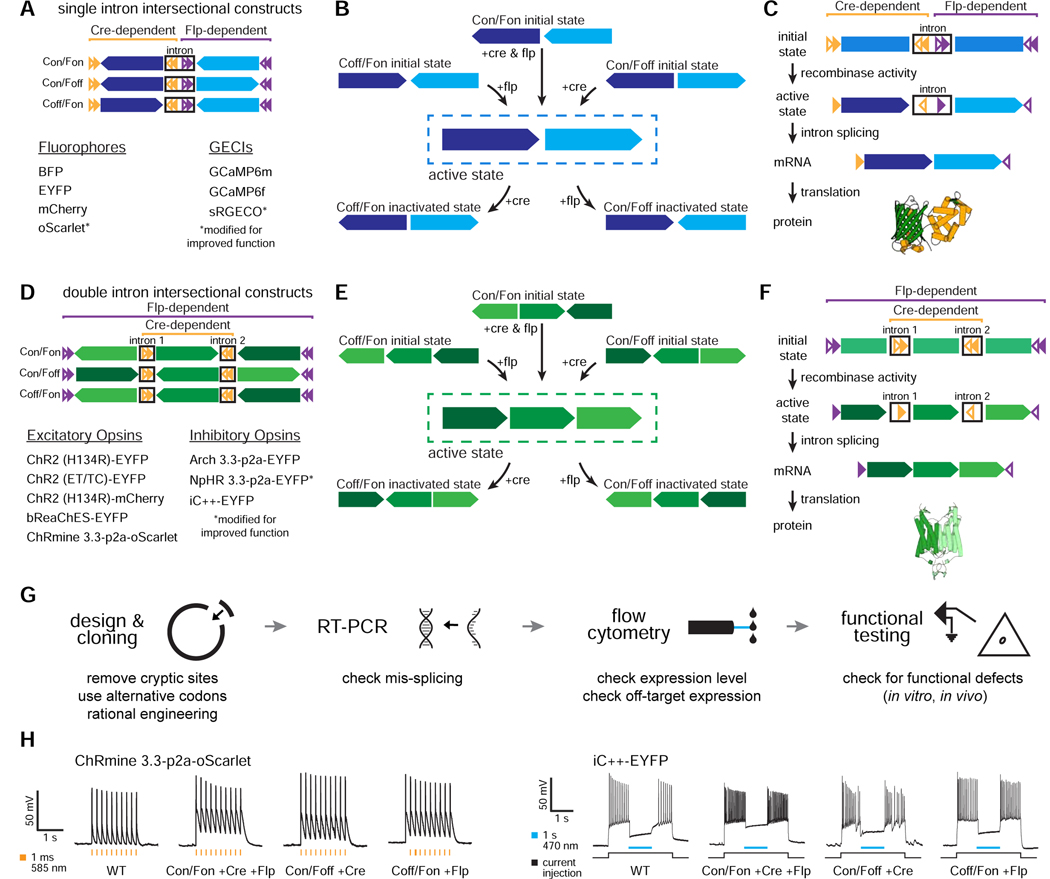
Figure 1. INTRSECT strategy and engineering pipeline.
A,D) INTRSECT molecular designs for single open reading frame (‘ORF’; A) and double ORF (D) in three boolean configurations (Cre AND Flp, Cre AND NOT Flp, Flp AND NOT Cre). Reagents for each configuration are listed. B,E) Activity of Cre and Flp to move the single ORF (B) and double ORF (E) INTRSECT starting configurations (top) to the active (dotted box, middle), and inactivated (bottom) states. C,F) From top to bottom, how initial DNA configuration for single ORF (C) and double ORF (F) constructs transition to the active state after recombinase-dependent rearrangement, mRNA processing that removes introns containing recombinase recognition sites, and translation without addition of extraneous sequence (crystal structure in C: GCaMP6m PDB: 3wld (Ding et al., 2014); F: iC++ PDB: 6csn (Kato et al., 2018)). G) Standardized engineering pipeline for production of novel INTRSECT constructs consisting of (left to right) design of intron placement and cloning, RT-PCR to ensure proper splicing, flow cytometry to assay proper expression, and functional testing (in cultured neurons or HEK cells) to compare with the parent. H) Electrophysiology in cultured neurons expressing WT, Con/Fon, Con/Foff, or Coff/Fon variants of ChRmine 3.3-p2a-oScarlet (left) or iC++-EYFP (right) with recombinases. See Figs. s1–s4.