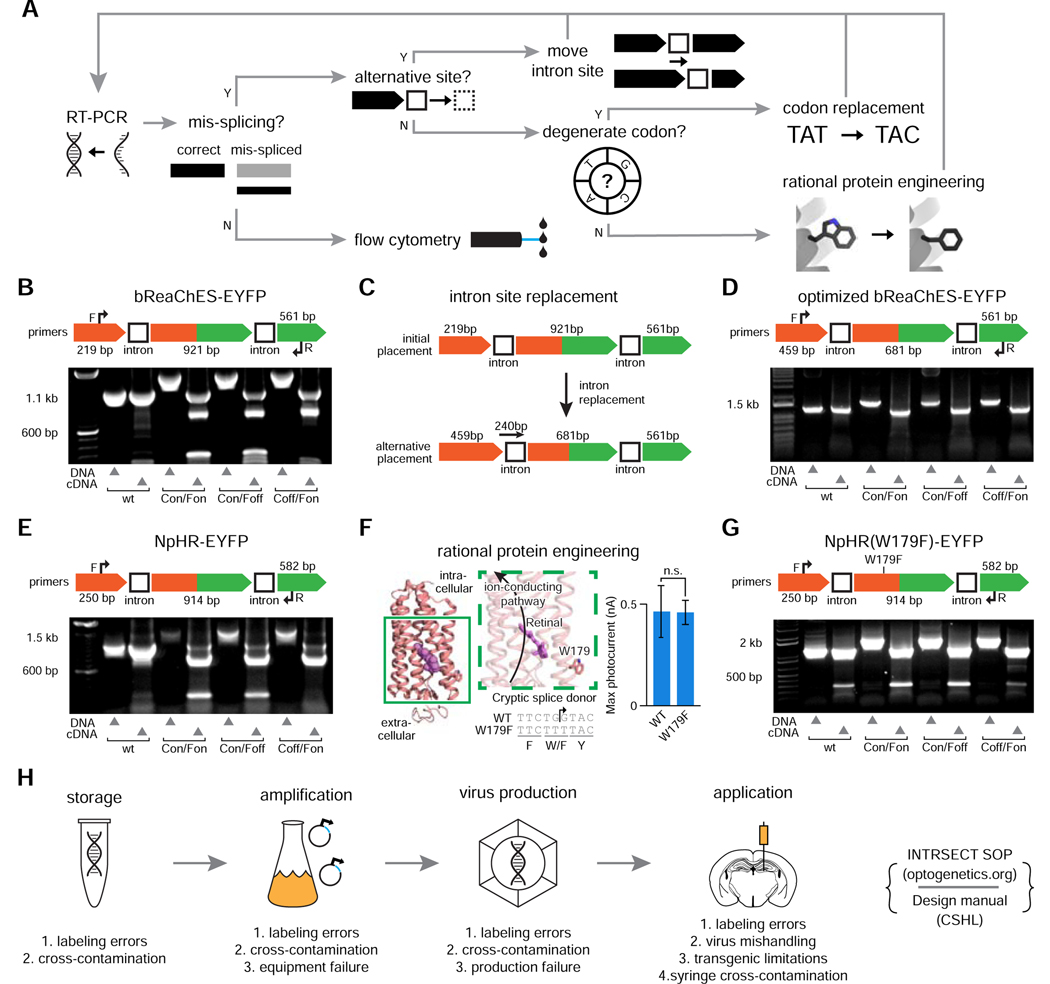
Figure 2. Standardized INTRSECT design/implementation.
A) RT-PCR testing and mis-splicing resolution approach for new INTRSECT constructs. B,E) Mis-spliced RT-PCR results for INTRSECT bReaChES-EYFP and NpHR3.3-p2a-EYFP. bReaChES-EYFP (B) and NpHR3.3-p2a-EYFP (E) had major and minor splice variants from cryptic splicing (noted by #) and exon1 to exon 3 direct splicing (noted by *). C,F) The bReaChES-EYFP intron was moved to an alternative splice site (C). NpHR3.3-p2a-EYFP did not have a separate splice site or degenerate codon options; guided by the published crystal structure, we disrupted the cryptic splice site (F-arrow) by introducing the mutation W179F (F-center), which did not affect opsin function (F-right; p = 0.9754, unpaired t-test). D,G) These iterations of bReaChES-EYFP (D) and NpHR3.3-p2a-EYFP (G) generated either single spliced products (D), or the correct major product and an exon 1-exon 3 minor splice variant (G). H) To catch errors early during scaling/implementation, we have described a protocol for making new INTRSECTs (Fenno et al., 2017) and maintain a Standard Operating Procedure (http://www.optogenetics.org/intrsect_sop.pdf). See Figures s1–s4.