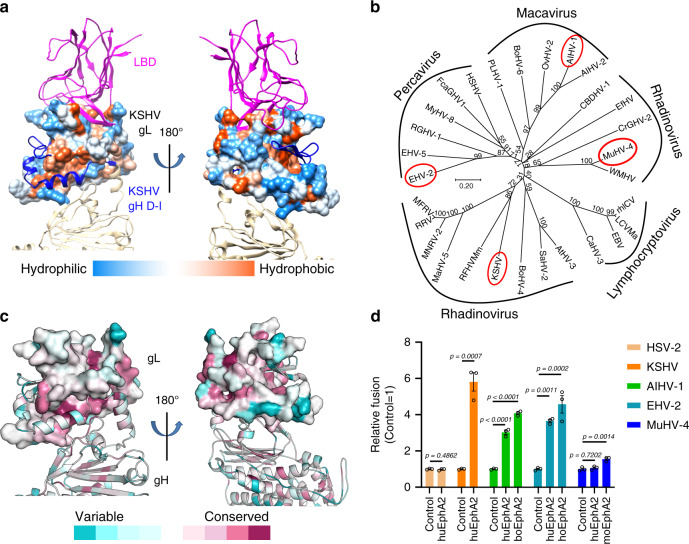
Fig. 6. EphA2 may serve as an entry receptor for multiple γ-herpesviruses.
a The structure of KSHV gL shows hydrophobic (orange) surfaces on both the EphA2 and the gH-binding faces. Molecular surfaces are colored according to hydrophobicity, with blue, white, and orange corresponding to the most hydrophilic, neutral, and hydrophobic patches, respectively. b Phylogenetic analysis of the gL proteins from 29 γ-herpesviruses, as generated by MEGA 10.146. Analyses were performed using the neighbor-joining method. The viruses selected for the cell fusion experiments are shown in red ellipses. c Conserved gL sequences mapped onto the protein surface. The gL surface is gradiently colored by the ConSurf server47, based on degree of conservation as indicated by the alignment of gL sequences from 29 γ-herpesviruses: the most conserved regions are dark magenta and the most divergent regions are dark cyan. d Cell-based fusion assays were performed by co-culturing of HEK-293T cells transfected with EphA2 and HEK-293T cells transfected with plasmids expressing viral gHgL and gB. The Macavirus AIHV-1, Percavirus EHV-2, and Rhadinovirus MuHV-4 were randomly selected for testing. KSHV was used as the positive control and HSV-2 was used as the negative control. Representative results from three experiments are shown. Relative fusion was normalized to the empty vector. The data are presented as mean ± SEM (n = 3 independent replicates). Unpaired Student’s t-test was used for comparing two groups and ordinary one-way ANOVA with Dunnett’s multiple comparison test for multiple comparisons. boEphA2: bovine EphA2; hoEphA2: horse EphA2; huEphA2: human EphA2; moEphA2: mouse EphA2. The GenBank accession codes of gH, gL, and gB are shown in the Supplementary Table 3. Source data are provided as a Source Data file.